Figure 1.
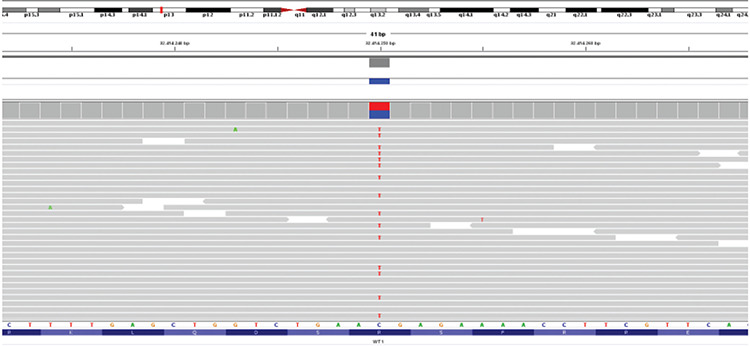
Next generation sequencing analysis WT1: variant visualization on integrative genome viewer (IGV). Patient DNA was sequenced using a custom panel including genes involved in 46,XX disorder of sex development. Sequence enrichment was performed using the NimbleGen SeqCap Target Enrichment kit (Roche) and sequenced on the Illumina NextSeq550 platform (Illumina, San Diego, California). VariantStudio software (Illumina, http://variantstudio.software.illumina.com/) was used for variants annotation. Each single variant has been evaluated for the coverage and the Qscore, and visualized via IGV software. The variant was analyzed in silico using prediction pathogenicity software (Scale-Invariant Feature Transform-SIFT and Polymorphism Phenotyping v2 -PolyPhen2) and database of variants frequency
