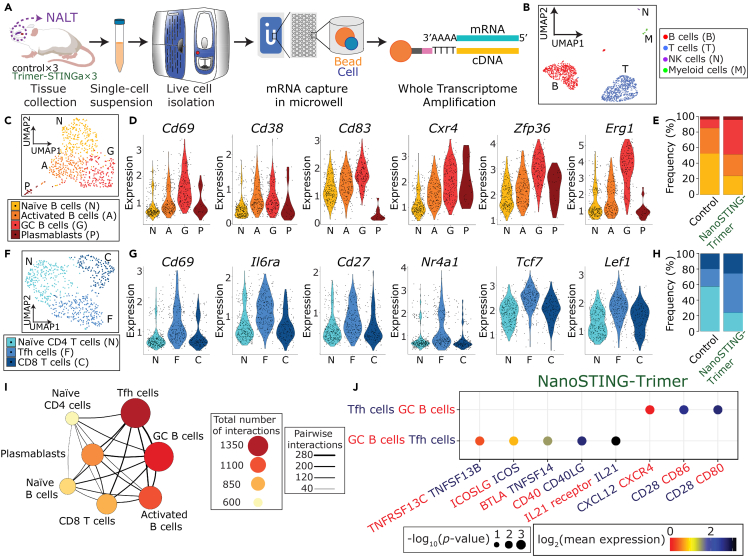
Figure 3.
scRNA-seq confirms the nasal-associated lymphoid tissue (NALT) as an inductive site
(A) Schematic of the experimental design for scRNA-seq on the NALT.
(B) Uniform manifold approximation and projection (UMAP) of the NALT immune cell profiles.
(C) Four clusters of B cells were identified based on UMAP: naive (N), activated (A), and germinal center (GC) B cells (G); and plasmablasts (P).
(D) Violin plots of the relative expression of Cd69, Cd38, Cd83, Cxcr4, Zfp36, and Erg1 in each of the four B-cell clusters.
(E) Bar plot illustrating the relative frequencies of each of the B-cell clusters in the NALT comparing the control and NanoSTING-Trimer groups.
(F) Three clusters of T cells were identified based on UMAP: naive (N) and follicular helper CD4 T cells (F); and CD8 T cells (C).
(G) Violin plots of the relative expression of Cd69, Il6ra, Cd27, Nr4a1, Tcf7, and Lef1 in each of the three T-cell clusters.
(H) Bar plot illustrating the relative frequencies of the different T-cell clusters in the NALT comparing the control and NanoSTING-Trimer groups.
(I) Cell-cell interaction network illustrating the interactions between the immune cells in the NALT. The size and color of the circles reflect interaction counts; large and darker circles indicate more interaction with other cell types. The thickness of the connecting lines reflects the relative number of interactions between each pair of cells.
(J) Predicted ligand-receptor interactions between the GC B cells and the Tfh cells within the NALT. The relevant ligand-receptor pairs are shown as bubble plots.
See also Figures S5–S8.