Figure 3.
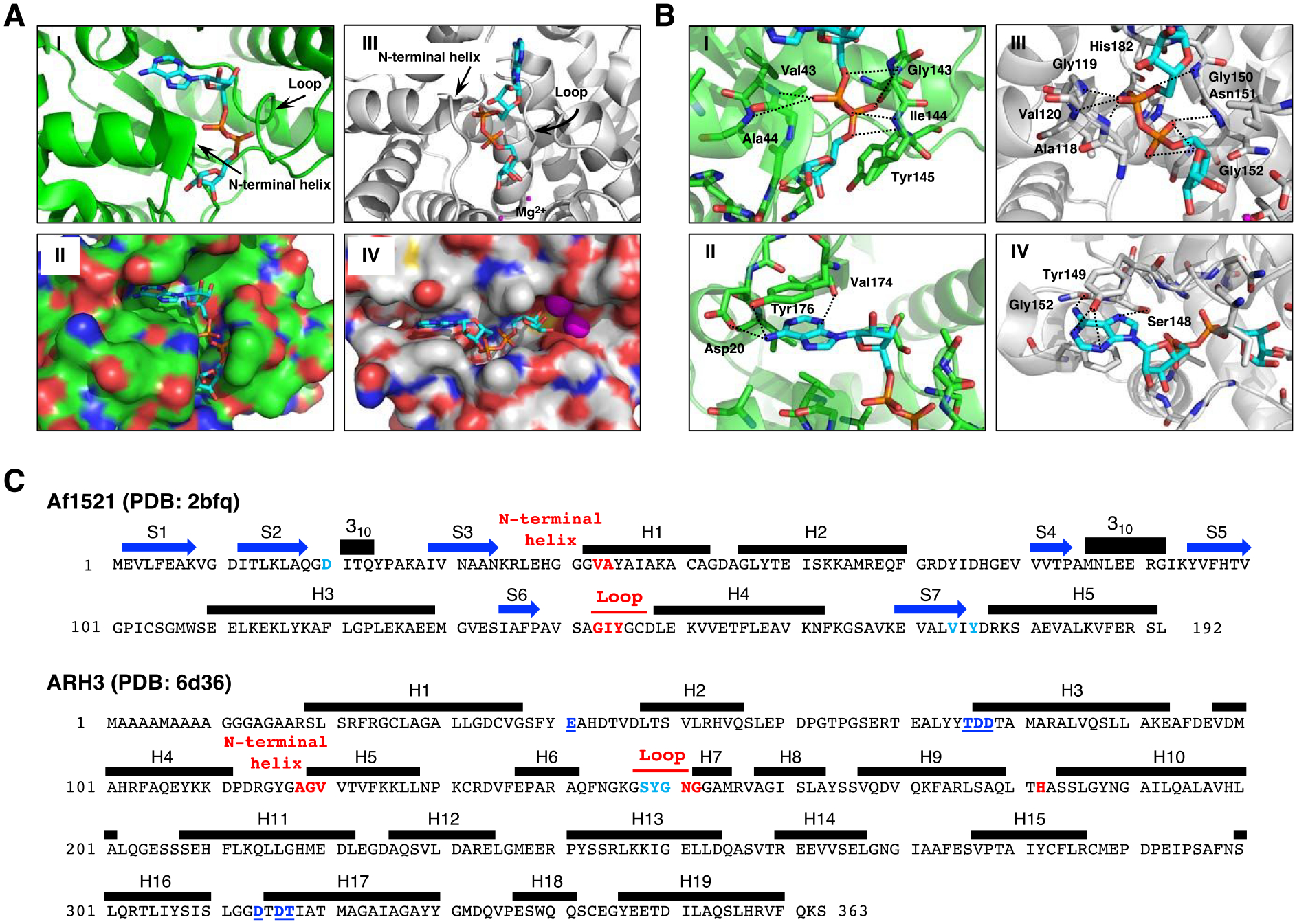
Structural Comparison of Af1521 (green) and ARH3 (gray) with bound ADP-ribose (cyan).
A. Crystal structure of Af1521 with bound ADP-ribose (cyan) (Macromolecular Structures Portal, PDB ID:2bfq). Arrows indicate the loop containing the diphosphate-binding motif, which is conserved in all macro domains39, 52; the N-terminal region of the adjacent alpha-helix is also involved in diphosphate binding (I). Surface representation of Af1521 with electro-negative and -positive atoms is shown respectively in red and blue. A binding-pocket for ADP-ribose is seen on the surface of Af1521 (II). Crystal structure53–55 of human ARH3 with ADP-ribose (PDB ID: 6d36). Arrows indicate the loop and N-terminal helix near the diphosphate group (III). Surface representation of ARH3 with electro-negative and -positive atoms is shown respectively in red and blue. A binding-pocket for ADP-ribose is also seen on the surface of ARH3 (IV)
B. Structural comparison of ADP-ribose coordination in the Af1521 macrodomain (green) and ARH3 (gray) pockets derived from Af1521, PDB ID:2bfq, and ARH3, PDB ID:6d36. Close-up view of Af1521 macrodomain (I) or ARH3 (III) in complex with ADP-ribose (cyan), focusing on the diphosphate of ADP-ribose in red. Residues near the distal diphosphate are shown in stick representation; hydrogen-bonding interactions are indicated with black-dashed lines. (II), (IV) Close-up view of Af1521 macrodomain (II) or ARH3 (IV) in complex with ADP-ribose, focusing on the adenine unit of ADP-ribose in blue. Residues near the adenine are shown in stick representation; hydrogen-bonding interactions are indicated with black-dashed lines.
C. Amino acid sequence and secondary structure elements of Af1521 (PDB ID:2bfq) and Human ARH3 (PDB ID:6d36). Based on the crystal structures depicted in Figure 3A,B (Supporting Information S10), the amino acids in Af1521 and hARH3 that interact with ADP-ribose and Mg2+, the diphosphate unit of ADP-ribose (red), the adenine unit of ADP-ribose (light blue) and hydrogen bonding residues near Mg2+ (blue with underline) are highlighted and colored. The N-terminal helix in Figure 3A corresponds to the segment indicated as H1 in Af1521 and H5 of hARH3. The amino acid sequence corresponding to the loop discussed in Figure 3A is also indicated in red. Blue arrows and black bars denote beta-strands (S) and alpha-helices (H), respectively.
