Figure 4.
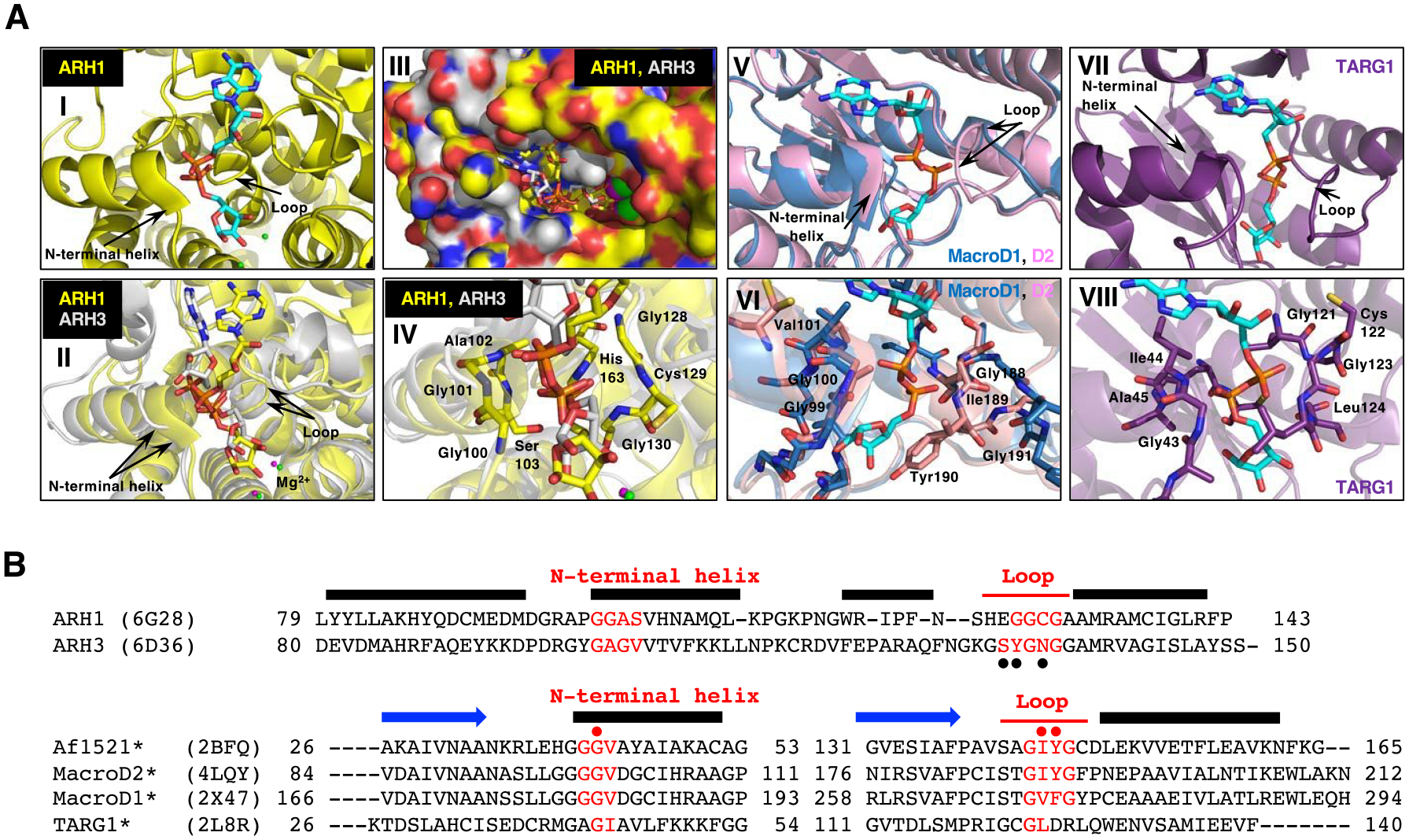
Structural comparison of macrodomain-containing proteins with bound ADP-ribose and ARHs with bound ADP-ribose.
A. Arrows indicate the loop and the N-terminal region of the adjacent alpha-helix that facilitates diphosphate binding in all these proteins with ADP-ribose (cyan) (I, II, V and VII). Overlay of the crystal structure of human ARH1 (yellow, PDB ID: 6g28) and ARH3 (gray, PDB ID: 6d36), with ADP-ribose (cyan) bound (II-IV). Overlay of the structure of the ADP-ribose-binding pockets in ARH1 and ARH3. Green (ARH1) and magenta (ARH3) small spheres indicate the bound Mg2+ ions (I and II) (Supporting Information S10). Overlay of ARH1 and ARH3 shown in a surface representation with a view of the ADP-ribose binding pocket (III). Close view of diphosphate unit of ADP-ribose (red) in ARH1 and ARH3. Residues of ARH1 near the diphosphate are highlighted (IV). Overlay of the crystal structures of human MacroD1 (blue, PDB ID: 2×47) and human MacroD2 (pink, PDB ID: 4lqy) with ADP-ribose bound (V). Crystal structure of human TARG1 (purple) with ADP-ribose bound (PDB ID: 2lbr) (VII).
Close-up view of MacroD1 and D2 macrodomain (VI, pink and blue) and TARG1 (VIII, purple) in complex with ADP-ribose (cyan), focusing on the diphosphate of ADP-ribose in red. Residues of MacroD2 and TARG1 near the distal diphosphate are shown in stick representation.
B. Structure-guided alignment of selected macrodomain proteins (lower) and ARHs (upper) protein sequence. The sequence regions corresponding to the N-terminal helix and loop that facilitate diphosphate binding with ADP-ribose are indicated (red). Secondary structure, N-terminal helix, loop and beta strand (blue arrows) elements are deduced from the Af1521, MacroD2, ARH1 and ARH3 crystal structures. Structure-guided alignment was generated with the ProMALS3D server. (•) amino acids involved in poly-ADP-ribose hydrolysis by ARH356, (• red) residues critical for catalytic activity (*) and selected macrodomain and ARH sequences protein for ADP-ribose binding properties48.
