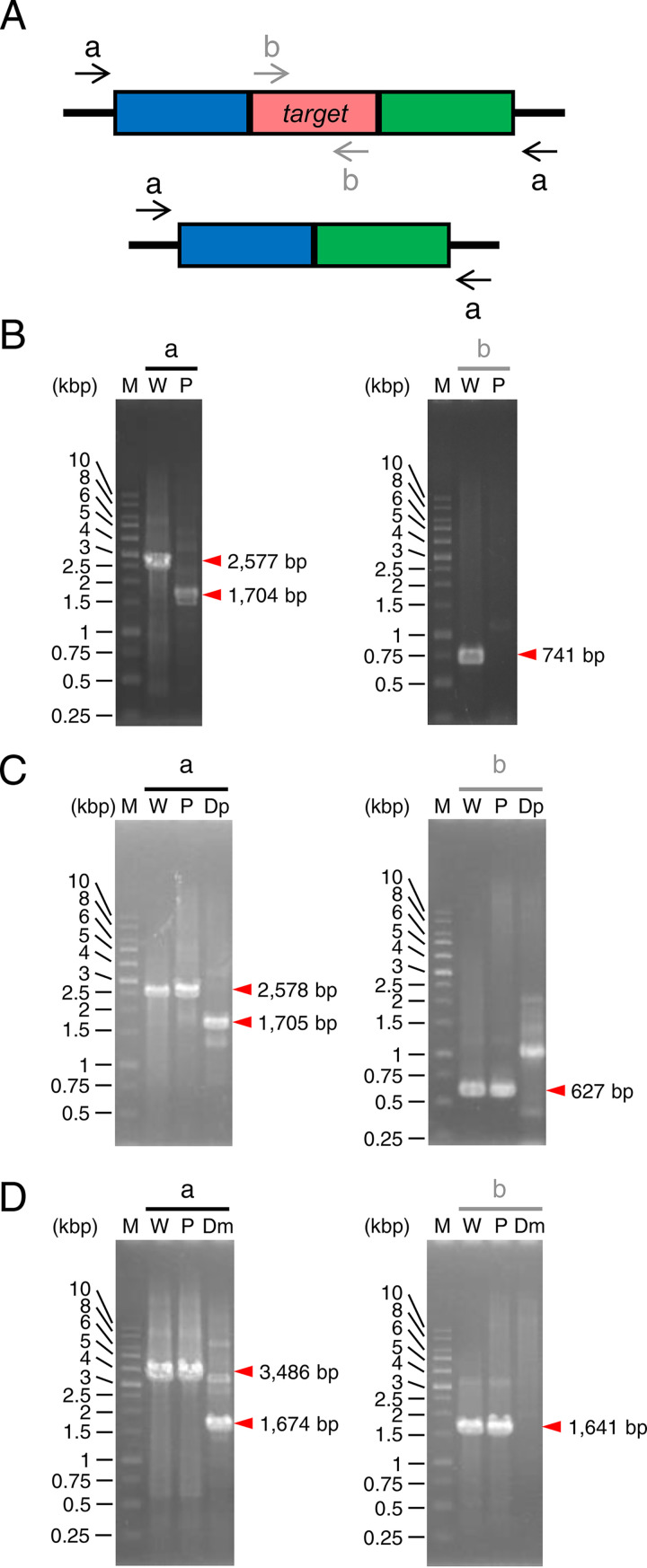
FIG 2.
PCR analyses of the gene disruption. (A) DNA primers that anneal to the genome before and after target gene disruption. These primers were used to analyze ΔpyrF (B), Δpetase (C), and Δmhetase (D) strains. The red box represents the disruption target region. The blue and green boxes represent its 5′- and 3′-flanking regions, respectively. The black arrows (a) and gray arrows (b) denote the primer set designed to anneal outside the target region for homologous recombination and within the target region, respectively. The expected number of base pairs in each DNA fragment is indicated at the right side of the red arrowhead. M, marker; W, wild-type strain; P, parent strain (ΔpyrF); Dp, Δpetase; Dm, Δmhetase.