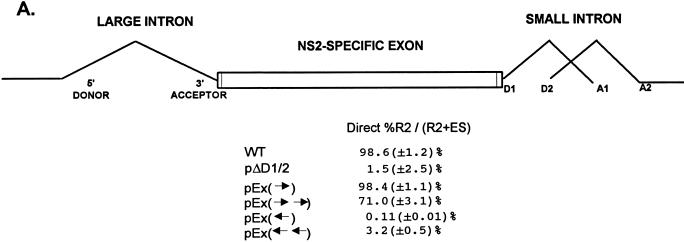
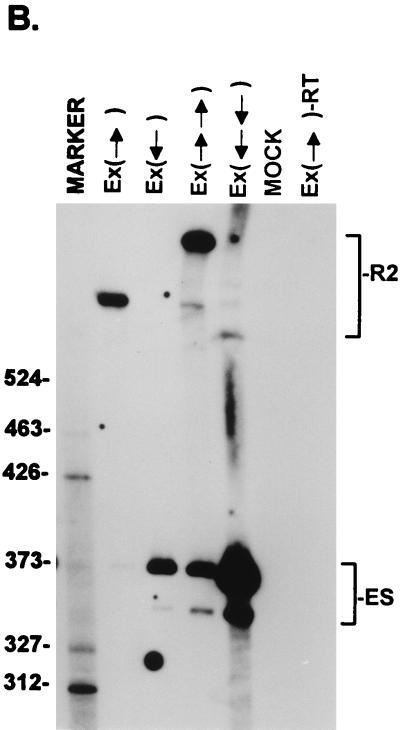
FIG. 7.
Inclusion of the NS2-specific exon tolerates doubling its length. (A) Quantitations of the direct percent R2/(R2+ES) ratio obtained by RT-PCR analysis for each mutant are shown. Construction of the mutants is explained in Materials and Methods. All the values are the average of at least three separate experiments. Standard deviations are indicated in parentheses. (B) RT-PCR analysis of RNA generated by wild-type MVM (WT), mutants (as described in the text), or mock transfected, as designated at the top of each lane. Samples were run on a 6% acrylamide–urea gel. MARKER is as described in the legend to Fig. 2B (the sizes of the marker bands are shown on the left). (Ex→)-RT is a control reaction with p(Ex→)-generated RNA but excluding reverse transcriptase. RNA generated by the wild type showed a 658-nt amplified R2 product, while RNA generated by the mutants showed R2 products of sizes consistent with the sizes of the substitutions in these mutants. As explained in the legend to Fig. 5D, two kinds of amplified ES, using either A1 or A2, were observed. The parent mutant bearing only the Sma sites at nt 2005 and 2270 [pEx(→)] is indistinguishable from the wild type with respect to splicing of the upstream intron and inclusion of the NS2-specific exon (56). Inclusion of the exon that is doubled in length still required wild-type exon sequences; when either one copy or two copies of the exon in the reverse orientation were inserted between the Sma sites [pEx(←) and pEx(←←), respectively (56)], the exon was almost uniformly skipped.