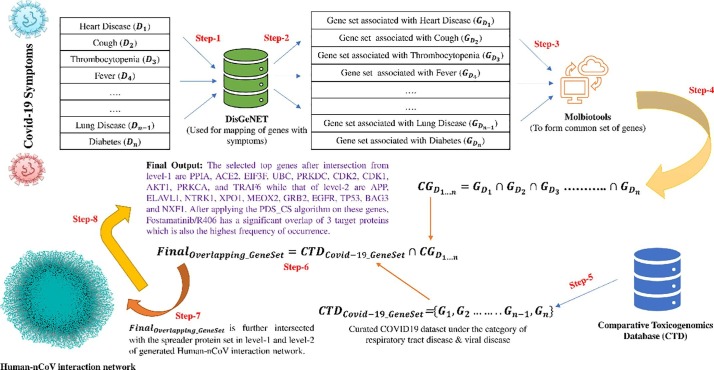
Fig. 3.
Covid19 Symptoms based analysis. The analysis consists of the following steps. Step-1: Symptoms are searched in the DisGeNET database. Step-2: Symptom associated gene sets are fetched from the DisGeNET database. Step-3: All gene sets are provided as an input to Molbiotools online. Step-4: Common overlapping set of genes are obtained from Molbiotools. Step-5: Curated Covid19 dataset is extracted from the Comparative Toxicogenomics Database (CTD) under respiratory tract disease & viral disease. Step-6: These sets of genes intersect with the common overlapping set of genes obtained from Molbiotools to form a key set of Covid19 related genes. Step-7: This key set of Covid19 related genes, after mapping to their corresponding protein IDs, are compared with the spreader proteins in Human-nCoV PPIN. Step-8: After comparison, top genes are selected in both level-1 and level-2 of Human-nCoV PPIN, which are finally used for the PDS_CS algorithm to determine the most potential candidate FDA drug for COVID19. Here, Fostamatinib/R406 has the highest DCS score.