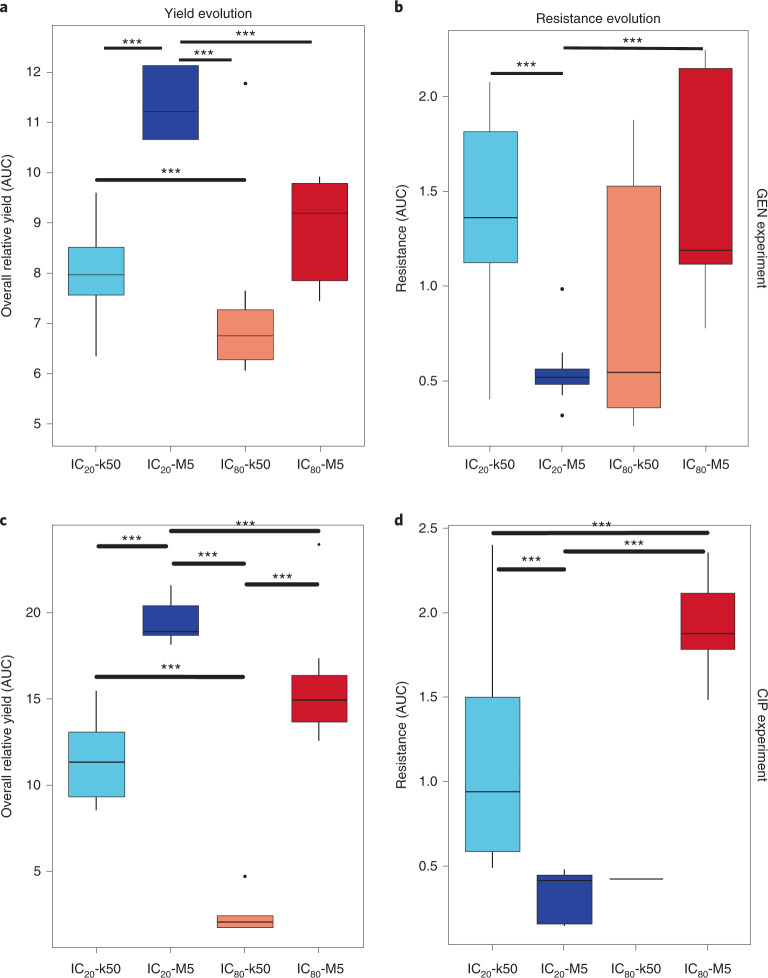
Fig. 1. Variation in bottleneck size and drug-induced selection leads to consistent responses across two independent evolution experiments with distinct antibiotics.
a,b, Results of the GEN experiment, showing changes in overall yield across experimental evolution relative to the no-drug control IC0 (a) and changes in overall resistance (b). c,d, Results of the CIP experiment, showing changes in overall yield across experimental evolution (c) and variation in resistance (d). The x-axis and colours represent different treatment groups (blue, IC20; red, IC80; light colours, 50k transfers; dark colours, 5 M transfers). For each replicate population, overall yield was inferred from counts of viable cells determined by flow cytometry at the end of each transfer period and related to the cell counts of the corresponding no-drug control of the same bottleneck size, followed by calculation of the AUC of yield across transfer periods of the evolution experiment. Resistance is shown as AUC of a standardized dose–response curve for the bacterial populations from the end of the evolution experiment (Methods and Extended Data Fig. 6). For the IC80-k50 treatment of the CIP experiment, only 1 out of 8 replicate populations survived, whereas in all other cases all 8 replicate populations per treatment were considered. In all box plots, which the centre line indicates the median, the box represents the data between first and third quantiles, the whiskers show the whole data range excluding outliers, and the dots indicate outliers (if present). Variation among treatments was evaluated with a general linear model; significant difference between two treatment groups, ***P < 0.05; Tukey’s honest significant difference (HSD) tests.