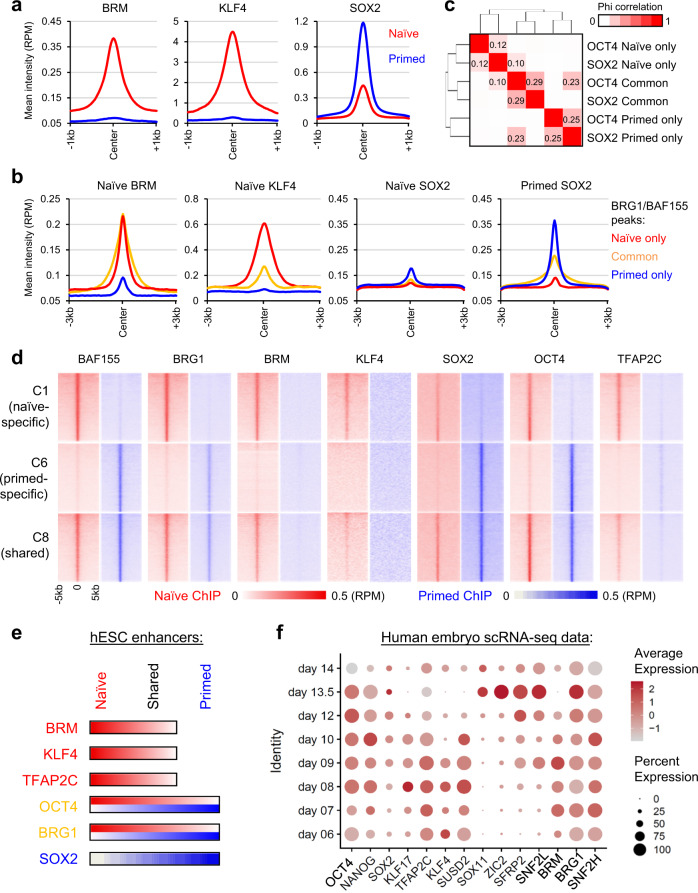
Fig. 4. Naïve and primed-specific enhancers harbor distinct chromatin remodelers and TFs.
a Mean intensity plots of BRM, SOX2 (ChIP-seq), and KLF4 (CUT&Tag) signals in naïve and primed hESCs. b Mean intensity plots of BRM, KLF4, and SOX2 signals at naïve-only, primed-only, and naïve/primed-common BAF peaks. c Correlation analysis of OCT4 and SOX2 chromatin association in naïve and primed hESCs grouped by naïve-only, primed-only, and naïve/primed-common OCT4 or SOX2 peaks. d Intensity heatmaps of BAF subunits, KLF4, SOX2, OCT4, and TFAP2C signals at clusters C1, C6, and C8 regions in naïve (left) and primed (right) hESCs. e Summary of activities of chromatin remodeling complexes and TFs in naïve (C1), primed (C6), and shared (C8) enhancers in hESCs. f Expression of OCT4-associated chromatin remodelers and TFs in single-cell RNA-seq analysis of human embryos cultured in vitro between days 6 and 14 of human development57. Average gene expression levels and the percentage of cells that express each gene are presented with differential color intensities and circle sizes, respectively. Note that KLF17 and SUSD2 are markers associated with naïve pluripotency, while SFRP2 and SOX11 are associated with primed pluripotency.