Fig. 4. Molecular evolution of macrolide and amikacin susceptibility within M. abscessus clone A3.
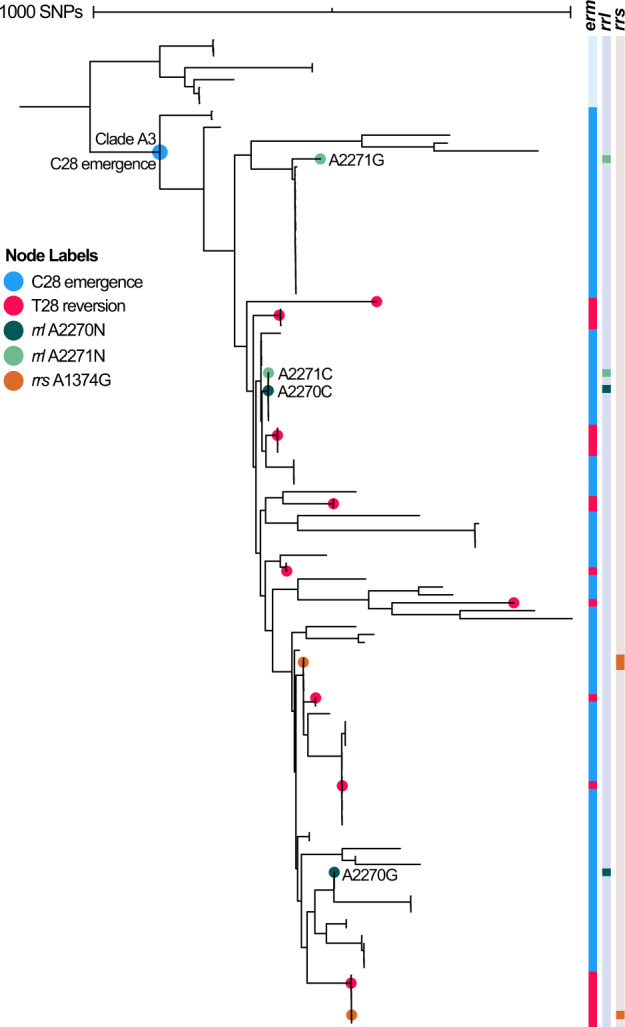
Midpoint rooted maximum-likelihood phylogeny of 116 members of clone A3. Each evolutionary gain or loss of a drug resistance mutation was assigned to its position on the phylogeny using parsimony. Given its basilar position on the phylogeny, the T28C erm(41) variant (blue circle), which precludes inducible macrolide resistance, occurred earlier in time, with subsequent-independent reversions back to T28 (red circles), indicating a regain of the inducible macrolide resistance phenotype. Within the clade, additional acquisitions of constitutive macrolide and amikacin resistance were also observed, with emergence of rrl A2270C (E. coli A2058C), rrl A2270G (E. coli A2058G), rrl A2271C (E. coli A2059C), rrl A2271G (E. coli A2059G), and rrs A1374G (E. coli A1408G). Bootstrap values (≥80%) support the independent, convergent acquisition of each instance of macrolide and amikacin resistance.
