Figure 4. Epigenetic modification preferences of mSWI/SNF complexes are defined by module-specific histone binding properties.
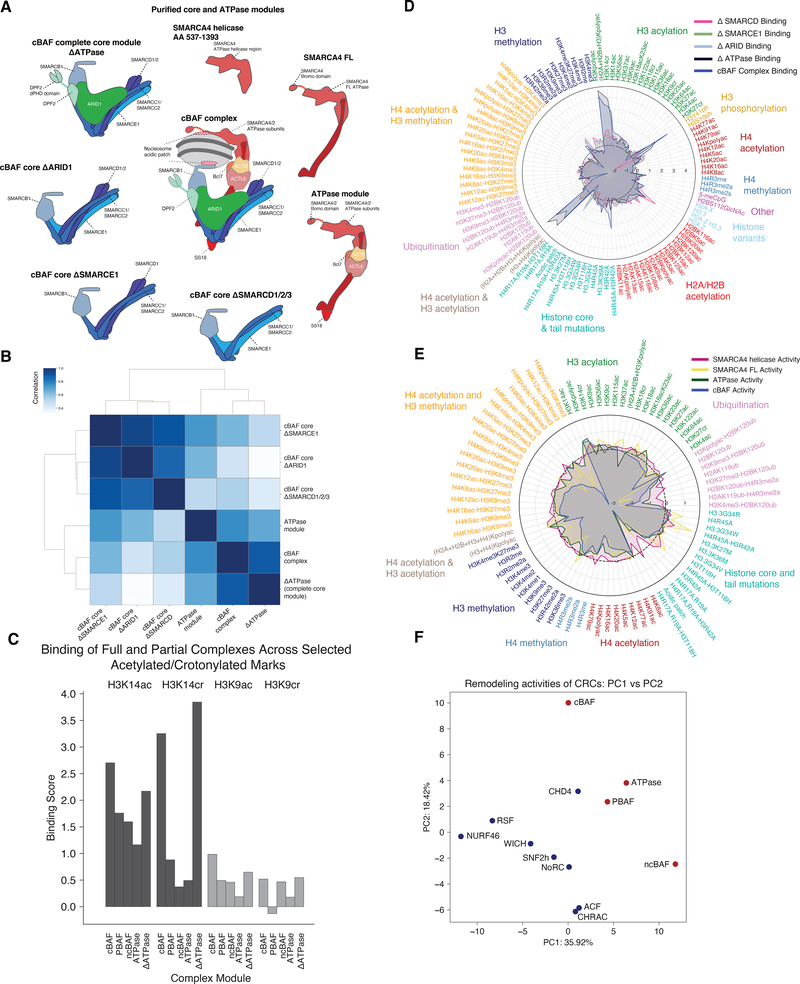
A. Schematic summarizing the cBAF core and ATPase modules/subunits subjected to full library binding and activity experiments.
B. Correlation heatmap for pan-library binding profiles for all cBAF core modules, ATPase module, and full cBAF complexes.
C. Binding scores for cBAF, PBAF, ncBAF complexes as well as the full core module (Core, ΔATPase) and the ATPase module (ATPase) over H3 lysine acylation marks (H3K14ac, H3K14cr, H3K9ac, and H3K9cr).
D. Radar plots indicating the binding of cBAF cores (ΔARID, ΔSMARCD, ΔSMARCE1, ΔATPase) and full cBAF complexes across all mononucleosomes profiled in the library. Marks and variants are separated by color. The radar plots are sorted by cBAF full complex binding within each histone mark type.
E. Radar plots indicating the remodeling activities of the ATPase module, SMARCA4 FL, truncated SMARCA4 (aa 537–1393) and full cBAF complexes across all mononucleosomes profiled in the library. Marks and variants are separated by color. The radar plots are sorted by cBAF full complex binding within each histone mark type.
G. Principal component analysis (PCA) of mSWI/SNF, CHD4 and ISWI complex activities.