Abstract
Long interspersed nuclear element-1 (L1) is a retrotransposable element that autonomously replicates in the human genome, resulting in DNA damage and genomic instability. Activation of L1 in senescent cells triggers a type I interferon response and age-associated inflammation. Two open reading frames encode an ORF1 protein functioning as messenger RNA chaperone and an ORF2 protein providing catalytic activities necessary for retrotransposition. No function has been identified for the conserved, disordered N-terminal region of ORF1. Using microscopy and NMR spectroscopy, we demonstrate that ORF1 forms liquid droplets in vitro in a salt-dependent manner and that interactions between its N-terminal region and coiled-coil domain are necessary for phase separation. Mutations disrupting blocks of charged residues within the N-terminus impair phase separation, whereas some mutations within the coiled-coil domain enhance phase separation. Demixing of the L1 particle from the cytosol may provide a mechanism to protect the L1 transcript from degradation.
Significance
Over half of the human genome is comprised of repetitive sequences. The long interspersed nuclear element-1 is an autonomous mobile DNA element that can alter its genomic location, resulting in genomic instability and DNA damage. Long interspersed nuclear element-1 encodes two proteins that are required for this function: the ORF1 RNA chaperone and the enzymatic ORF2. Here, we demonstrate that ORF1 forms liquid-liquid phase-separated states in vitro, mediated by electrostatic interactions between the conserved, disordered N-terminus and coiled-coil domain of ORF1.
Introduction
Over half of the human genome is composed of repetitive sequences (1), the largest fraction of which are mobile DNA sequences known as retrotransposable elements (RTEs) (2). They are comprised of three major classes, the long terminal repeat (LTR) elements (related to retroviruses) and the non-LTR elements, known as long interspersed nuclear elements (LINEs) and short interspersed nuclear elements. RTEs colonize their host genomes using a “copy-and-paste” mechanism that employs an RNA intermediate and an RTE-encoded reverse transcriptase (RT) enzyme (3). RTEs can exert deleterious effects on their host cells in many ways, including insertional mutagenesis, promotion of DNA damage and genomic structural instability, triggering of epigenetic changes, and disruption of normal patterns of gene regulation (4). The only autonomously active human RTEs are the LINE-1 elements (L1). L1s comprise ∼17% of the human genome (∼500,000 copies), but only the most evolutionarily recent subfamily L1HS has active members (5,6). Until recently, L1s were thought to be silent in somatic cells, but new evidence points to activity in the brain (7), cancers (8), cellular senescence (9), and aging (10, 11, 12). In senescent cells, transcriptional activation of L1 and the ensuing reverse transcription to its complementary DNA triggers a type I interferon response, which promotes age-associated inflammation, also known as inflammaging (11,12). Given that inflammaging has been linked with several important age-related, chronic diseases (13), further understanding of the details of the L1 life cycle in somatic cells is expected to provide novel therapeutic targets.
L1 is a 6 kb element whose 5′ untranslated region contains an internal promoter from which two proteins required for retrotransposition are expressed. The 40 kDa ORF1 protein functions as a messenger RNA (mRNA) chaperone and packing protein, whereas the 150 kDa ORF2 protein encodes the endonuclease and RT activities necessary for retrotransposition (Fig. 1 A; (3,14)). The L1 ribonucleoprotein (RNP) particle assembles in cis in the cytoplasm before entering the nucleus for integration into a novel site in the host genome (15). ORF2 binds the L1 transcript at its 3′ end, whereas the ORF1 protein coats the remainder of the mRNA (16). Retrotransposition in the nucleus occurs by a mechanism known as target-primed reverse transcription, which is initiated by an endonuclease-generated nick in the genome (2). The displaced flap of DNA binds to the poly A tail of the mRNA template and is then reverse transcribed at the site of insertion. To enable this life cycle, it is necessary to maintain an intact, full-length transcript for successful target-primed reverse transcription, which highlights the importance of the ORF1 chaperone functions (14,17).
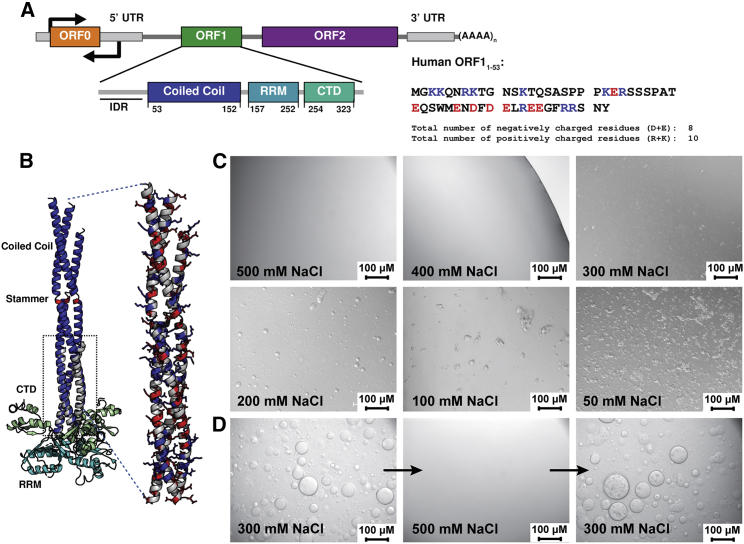
Figure 1.
ORF1 phase separation is driven by electrostatics and is reversible. (A) Domain organization of L1 and ORF1 functional units. ORF1 consists of three folded domains: a coiled-coil region responsible for trimerization, an RRM, and a CTD, which act in concert to bind to L1 RNA. The intrinsically disordered N-terminal region is denoted as IDR. The sequence of the N-terminal 53 residues is shown color coded by charge. (B) Composite model of the ORF1 trimer. The extended coiled-coil domain (Protein Data Bank entry PDB: 6FIA) is superposed on the ORF1 core structure (PDB: 2YKO) in the boxed region. Domains are colored as shown in (A). The stammer region is highlighted in red. An enlarged view of the coiled-coil domain shows the charge distribution on the solvent-exposed surface, with basic residues shown in blue and acidic residues in red. (C) The extent and morphology of the ORF1 phase-separated state is dependent on the NaCl concentration in the buffer solution. In low salt concentrations, ORF1 forms amorphous aggregates while forming spherical droplets in ranges of 200–400 mM NaCl. In 500 mM NaCl or higher, ORF1 remains dispersed. All samples were imaged at room temperature in 20 mM Tris (pH 8.0) and 1 mM DTT with the noted NaCl concentration and 50 μM ORF11–338. (D) ORF1 phase separation is reversible. Increasing the NaCl concentration from 300 to 500 mM while maintaining protein concentration (300 μM) solubilizes phase-separated ORF1. Phase-separated droplets reappear when the NaCl concentration is reduced to 300 mM. Data were collected at room temperature in 20 mM Tris (pH 8.0) and 1 mM DTT in either 300 or 500 mM NaCl as noted on the micrograph. To see this figure in color, go online.
Two crystal structures of the ORF1 protein have been determined and indicate that ORF1 trimerizes via its coiled-coil domain (Fig. 1 B; (18,19)). The C-terminal portion of the protein contains an RNA recognition motif (RRM) and the C-terminal domain (CTD). Together, these two domains form a positively charged surface that is predicted to weave the single-stranded L1 RNA around the base of the ORF1 trimer (18,19).
The 150 Å-long coiled-coil domain is organized in a series of 14 heptads with small hydrophobic residues in positions two and four that orient to the center of the helical bundle formed by the trimer. The C-terminus of the coiled coil contains seven heptads with repeating R-h-x-x-h-E motifs (where x denotes any amino acid and h denotes the hydrophobic residues at positions 2 and 4) that form salt bridges to stabilize trimerization. This sequence is highly conserved across species (18). Residues 91–93 (MEL) are inserted within heptad 9 and form a three-residue bulge, or stammer, that destabilizes the N-terminal portion of the coil and contributes to an observed metastability that seems to be required for L1 retrotransposition (Fig. 1 B; (19)). This suggests that the N-terminal portion of the coil can switch between fully structured or partially unstructured states, which is corroborated by lacking electron density for heptads 13–14 within the shortest helix in the crystal structure (19). Despite the fact that ORF1 has been well characterized structurally, less is known about the function of the N-terminal region. Mutations in ORF1 that disrupt nucleic acid binding, deletion of the stammer insertion in the coiled-coil domain, or removal of the positive charge at the N-terminus all drastically impair retrotransposition to an extent similar to abrogating RT activity (18, 19, 20, 21). In addition, the expression of truncated ORF1 proteins containing the N-terminal and coiled-coil region suppresses L1 retrotransposition, supporting the importance of these domains for L1 function (22).
In cell culture overexpression systems, the ORF1 protein has been observed within two membrane-less organelles, stress granules and processing bodies (P-bodies), in the cytoplasm of cells, both in the presence and absence of cellular stress (23,24). Tandem affinity capture of fully assembled cytoplasmic L1 RNP particles, using a FLAG antibody against tagged ORF2 followed by a monoclonal ORF1 antibody, identified interactions between several stress granule or P-body-associated proteins and the fully assembled cytoplasmic L1 RNP (25, 26, 27, 28). It is possible that stress granules provide an additional layer of protection from circulating proteases, RNases, and immune sensors and give the L1 RNP complex additional time to properly form. This phenomenon has been observed for the assembly of the LTR retrotransposon Ty3 into a capsid-like structure in yeast P-bodies (29). On the other hand, sequestration within stress granules may target the L1 RNP assembly for degradation as an additional host defense response against retrotransposition (23).
The formation of stress granules and other membrane-less organelles can be driven by liquid-liquid phase separation (LLPS), often initiated by RNPs, to partition the cellular space and exert spatiotemporal control over biochemical reactions within each particle (30,31). Weak intermolecular interactions between proteins, nucleic acids, and combinations thereof drive coalescence into a phase-separated state where biomolecules coexist in condensed and dilute phases. In the condensed phase, these biomolecules are at high local concentrations that function to exclude solvent and other biomolecules in favor of interactions with specific proteins or nucleic acids (32). This forms a chemically distinct environment that is able to reversibly demix from the surrounding cytoplasm or nucleoplasm (33). Because liquid phase-separated states internally consist of highly dynamic interactions and lack a physical membrane barrier, some of these structures are able to rapidly form or disassemble in response to the cellular environment, exemplified by stress granule formation and disassembly during times of heightened cellular stress and the subsequent removal of the stressor (32). The ability of proteins to incorporate into coacervates is often encoded in modular domains that enable multivalent interactions or in intrinsically disordered regions that form complexes through intermolecular contacts in a sequence-dependent manner (30,34). For example, several proteins known to localize to physiological granules and that readily undergo phase separation in vitro, such as fused in sarcoma (35) or TAR DNA-binding protein-43 (TDP-43) (36), have low-complexity sequences, which are regions composed of short repeat sequences that mediate nonspecific interactions with other biomolecules within the phase-separated body. Furthermore, folded RNA-binding domains as well as disordered segments with arginine-rich sequences are features of both these proteins, contributing to multivalency of interaction (36). Although the protein does not contain easily identifiable repeat sequences, many of the critical features of phase-separating proteins are present in ORF1.
Because of the association of ORF1 with cytoplasmic phase-separated bodies, the requirement of its putatively disordered N-terminal domain for L1 retrotransposition, and the multivalent RNA-binding architecture similar to known phase-separating proteins, we hypothesized that the disordered N-terminal region promotes phase separation of the ORF1 protein. Using biophysical phase separation assays and NMR spectroscopy, we show that, although the presence of the N-terminus is necessary for ORF1 condensation, in vitro LLPS is also mediated by the interaction of the disordered N-terminus with the C-terminal portion of the trimeric coiled-coil domain. Interactions that drive ORF1 phase separation are dependent on electrostatics and can be modulated by the solution conditions and by mutations within the ORF1 protein.
Materials and methods
Cloning and expression of recombinant proteins
A plasmid containing the bacterial codon-optimized sequence of ORF11–338 was synthesized by GenScript in the pUC57 cloning vector (UniProt: Q9UN81, Table S1). ORF1 sequences of interest were PCR amplified using either Thermo Fisher Scientific Phusion Hot Start II High-Fidelity PCR Master Mix or ACTGene ACTaq High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA). The inserts were cloned into the pET26b vector for expression (Novagen, Madison, WI) using NdeI and XhoI double digested products for T4 ligation or undigested insert for Gibson assembly into the pET26b expression vector (37). ORF1 mutants were generated using the pET26b-ORF11–338 as a template for overlap extension PCR, as described previously (38). The ORF1 insert was PCR amplified with primers containing the mutation of interest and T7 promoter or terminator primers. The PCR product was gel extracted, and the forward and reverse products were mixed as a template for the next round of PCR amplification using the universal T7 promoter and terminator primers. The PCR product was gel extracted and cloned into an empty pET26b vector as described above.
ORF1 expression plasmids were transformed into Escherichia coli BL21STAR(DE3) cells (Invitrogen, Carlsbad, CA). One-liter cultures supplemented with 50 μg/ml kanamycin were inoculated with 20 ml of an overnight culture and grown at 37°C to an OD600 between 0.5 and 0.8. Cultures were cooled for 1 h to 20°C, and 0.5 mM isopropyl β-D-1-thiogalactopyranoside was added to induce protein expression for 16–18 h. Cells were harvested in a Beckman SLC-6000 rotor (Beckman Coulter, Brea, CA) at 5500 × g for 12 min at 4°C.
ORF1 purification
Cells were resuspended in 10 ml of lysis buffer (20 mM Tris (pH 8.0), 1 M NaCl, and 5 mM imidazole supplemented with phenylmethylsulfonyl fluoride and benzamidine) per 1 g of dry cell pellet mass and lysed using an Avestin EmulsiFlex C3 (ATA Scientific, Ottawa, Ontario, Canada). Lysate was clarified at 30,000 rpm for 60 min at 4°C in a Beckman Ti45 ultracentrifuge rotor (Beckman Coulter) and filtered through a 0.22-μm polyethersulfone membrane before being loaded onto a XK 16/20 column (Cytiva, Marlborough, MA) packed with 25 ml of QIAGEN Ni-NTA resin (QIAGEN, Hilden, Germany). The column was washed with a minimum of five column volumes of HisTag Buffer A (20 mM Tris (pH 8.0), 1 M NaCl, and 5 mM imidazole) and eluted with a gradient of 0–70% HisTag Buffer B (20 mM Tris (pH 8.0), 1 M NaCl, and 500 mM imidazole). Fractions corresponding to the ORF1 protein were pooled and concentrated to 5 mL using the Millipore 10 kDa Amicon Ultra Centrifugal Filters (MilliporeSigma, Burlington, MA) and injected onto a GE HiPrep 16/60 Sephacryl S-300 HR gel filtration column (Cytiva) equilibrated in 20 mM Tris (pH 8.0), 1 M NaCl, and 1 mM dithiothreitol (DTT). All constructs and mutants of ORF1 were purified using the same protocol, except ORF11–53, which was heat purified before gel filtration. Once concentrated to 5 mL, ORF11–53 was incubated at 70°C for 15 min. The protein was cooled on ice for 15 min, and any precipitation was removed by centrifugation at 12,000 rpm for 10 min at room temperature.
Microscopy
Samples of ORF1 for differential contrast (DIC) microscopy were prepared by diluting a 1–2 mM concentrated stock of the ORF1 protein into a lower salt buffer. 5 μL of the sample was spotted onto a glass coverslip for imaging using 20× magnification and Nomarski optics on a ZEISS Axiovert 200 M microscope (ZEISS, Jena, Germany). All microscopy images shown are of 300 μM ORF1 protein in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM DTT, unless otherwise noted.
For the titration experiments between ORF11–53 and either the full-length ORF11–338 or coiled-coil domain ORF153–152, either 2 mM (1:1), 6 mM (1:3), or 12 mM (1:6) ORF11–53 was added to 2 mM ORF153–152 or ORF11–338 in 20 mM Tris (pH 8.0), 1 M NaCl, and 1 mM DTT. All samples remained clear upon mixing. They were subsequently diluted to 300, 150, or 50 μM ORF153–152 or ORF11–338 in 500, 300, and 150 mM NaCl buffers. Phase separation was assayed by microscopy, as described above.
Dilute phase diagram
ORF1 was diluted to 300 μM in a final buffer of 20 mM Tris (pH 8.0) and 1 mM DTT with salt concentrations of either 100, 150, 200, 250, 300, 350, 400, 450, or 500 mM NaCl. 100 μL samples were incubated at 25°C in a thermocycler for 30 min. The condensed phase was separated from the dilute phase by centrifugation at 10,000 rpm for 30 s. Protein concentrations in samples of the dilute phase were measured directly on a Thermo Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific) using the appropriate buffer as a blank. Concentrations were calculated using the Beer-Lambert law with the predicted extinction coefficients and molecular weights corresponding to each construct. Errors bars denote the standard deviation of three technical replicates.
NMR spectroscopy
All NMR experiments were carried out at 25°C on Bruker Avance III HD 850 MHz or NEO 600 MHz spectrometers equipped with 5 mm TCI CryoProbes (Bruker, Billerica, MA) and single-axis pulsed-field gradients. NMR data were acquired on isotope-enriched ORF11–53 samples (20 μM to 1.6 mM) dissolved either in 20 mM HEPES (pH 7.25) or 20 mM MES (pH 6.0) buffers containing 200 mM NaCl, 1 mM DTT, and 5% D2O. NMR data were also acquired on isotope-enriched ORF11–152 samples at the specified protein concentrations in 20 mM MES (pH 6.0) buffer containing 400–933 mM NaCl, 1 mM DTT, and 7% D2O in 3-mm NMR tubes. All data were acquired using standard pulse programs provided by the instrument manufacturer. NMR data sets were acquired and processed using Topspin software version 3.5 or 4.0 (Bruker) and were further analyzed in Sparky version 3.112 (University of California, San Francisco). Backbone resonance assignments for ORF11–53 were achieved through the analysis of standard three-dimensional triple resonance experiments HNCA, HNCACB, and HNCO. 15N-relaxation experiments were performed on 15N-labeled ORF11–53 or in the presence of equimolar complex with nonlabeled ORF153–152. The values of the 15N-longitudinal relaxation rate constants were measured using interleaved experiments with relaxation delays of 20, 40, 60, 100, 200, 400, and 800 ms. The values of the transverse relaxation rate constants (R2) were measured using interleaved experiments with relaxation delays of 15.7, 31.4, 78.5, 110, 157, 173, and 251 ms. Both rate constants were calculated assuming a monoexponential decay of the peak intensities using the program Curvefit (Prof. Arthur Palmer, Columbia University). The steady-state heteronuclear {1H}–15N NOE experiment was carried out in an interleaved manner, with and without proton saturation, and the NOE effect was calculated as a ratio of the peak intensities.
Results
The full-length ORF1 protein phase separates and this property is dependent on electrostatic interactions
During purification of the recombinant full-length ORF11–338 protein, we observed that protein solubility is strongly dependent on salt concentration and that protein purification requires high salt concentrations to prevent protein aggregation. In gel filtration experiments of soluble ORF11–338 in 1 M NaCl buffer, the protein elutes at a position consistent with trimer formation via the coiled-coil domain, as previously described in structural studies of both an RNA-binding construct ORF1110–321 and an ORF153–152 construct that isolates the extended coiled-coil domain (Fig. S1; (18,19)). We have found that reduction of NaCl concentration promotes LLPS of full-length ORF11–338, and the extent of ORF1 condensate formation is dependent on both NaCl and protein concentrations (Figs. 1 C and S2). In buffers with 300 mM NaCl, ORF1 condensates form spherical droplets that behave as dynamic liquids, capable of fusing with neighboring droplets (Fig. S3).
By increasing the NaCl concentration to 500 mM while maintaining a 300 μM ORF1 protein concentration, ORF1 condensates are dispersed into a single phase with the protein evenly distributed in solution. Reducing the NaCl concentration back to 300 mM in the same sample subsequently reinitiates ORF1 condensation, demonstrating reversibility (Fig. 1 D). However, reducing the salt concentration to 50–150 mM NaCl causes the ORF1 protein to form irregularly shaped structures that do not fuse and flow, indicating that the ORF1 phase-separated state is capable of forming either higher-order structures or hydrogels. Similar amorphous structures can be observed in higher NaCl concentrations with increasing ORF11–338 concentration, such as 150 μM ORF11–338 at 200 mM NaCl (Fig. S4), indicating a critical threshold for the formation of higher-ordered structures within the spectrum of the phase-separated state. At NaCl concentrations between 500 mM and 1 M NaCl, ORF1 remains well mixed with the surrounding buffer. The observed salt dependency suggests that electrostatic interactions likely play an important role in biomolecular condensation of ORF11–338.
Truncated ORF11–152 containing the N-terminus and coiled-coil domain phase separates readily, with characteristics similar to full-length ORF1
To identify the domains involved in ORF1 phase separation, we designed constructs that isolated either individual or tandem domains. We found that the RRM-CTD tandem construct does not phase separate (Figs. 2 A and S5), whereas truncation of the N-terminal 53 or 65 residues greatly reduces the extent of phase separation (Fig. S6). In contrast, a minimal construct containing the N-terminus and coiled-coil domain (ORF11–152) phase separates readily, similar to the full-length protein, and displays analogous patterns of phase separation at lower salt concentrations as observed by DIC microscopy (Figs. 2 A and S7; Table 1). Therefore, we chose to analyze molecular interactions driving ORF11–152 phase separation in greater detail.
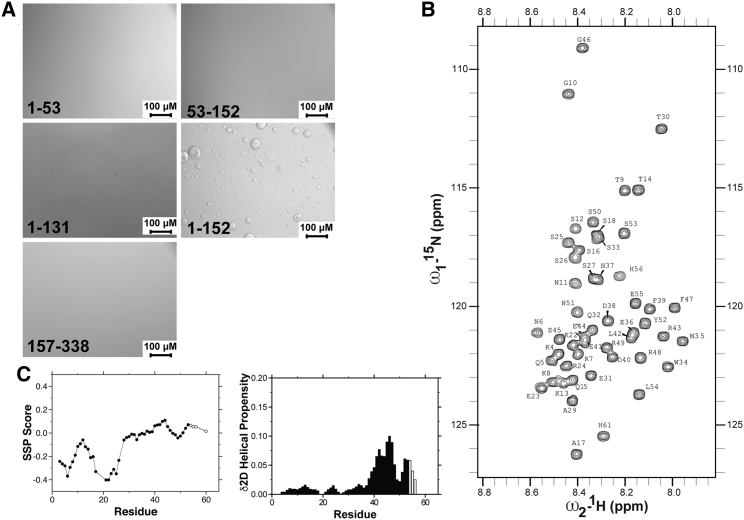
Figure 2.
ORF11–152 readily phase separates, and the N-terminal domain and coiled-coil domain are both required for phase separation. (A) Constructs of the N-terminal region (1–53), the coiled-coil domain (53–152), and truncation of the coiled-coil domain (1–131) in a tandem construct remain soluble at a lower salt concentration (300 mM NaCl). In contrast, the full-length tandem construct lacking the RRM and CTD (1–152) phase separates to the same extent as the full-length protein, indicating that both the N-terminus and coiled-coil domains are necessary for phase separation. All data were collected with 300 μM ORF1 at room temperature in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM DTT. (B) The narrow chemical shifts of ORF11–53 in the 15N-1H HSQC experiment demonstrate that this region is intrinsically disordered. Resonances beyond 53 correspond to the histidine tag. (C) The secondary structure propensity (SSP (39)) score and δ2D-helical propensity scores (40) show that residues 41–49 have slight α-helical character. Open circles and open bars correspond to residues from the histidine tag.
Table 1.
Summary of constructs used
| ORF1 construct | Domain organization | Phase separation in comparison with wild-type ORF11–338 | Retrotransposition | κ-Value (41) |
|---|---|---|---|---|
| ORF11–338 | full length | comparable | normal | 0.149 |
| ORF11–53 | N-terminal region | none | not determined | – |
| ORF11–152 | N-terminus + coiled coil | comparable | not determined | – |
| ORF153–152 | coiled coil | none | not determined | – |
| ORF153–338 | ΔN-terminus | reduced | none (19) | – |
| ORF166–338 | ΔN-terminus + truncated coiled coil | reduced | none (19) | – |
| ORF1157–338 | RRM + CTD | none | not determined | – |
| ORF1–338 K3A/K4A | full length | comparable | not determined | K3A = 0.147 |
| ORF1–338 G2A/K3A/K4A: none (42) | K4A = 0.146 | |||
| ORF11–338 K3R: increased (19) | K3A/K4A = 0.146 | |||
| ORF1–338 K3E/K4E | full length | reduced | not determined | K3E = 0.144 |
| K4E = 0.142 | ||||
| K3E/K4E = 0.144 | ||||
| ORF1–338 K3A/K4A/R7A/K8A | full length | reduced | not determined | 0.143 |
| ORF1–338 K3E/K4E/R7E/K8E | full length | reduced | not determined | 0.151 |
| ORF1–338 R7E/K8E | full length | reduced | not determined | 0.143 |
| ORF1–338 S27D | full length | increased | increased (43) | 0.150 |
| ORF1–338 L93P | full length | increased | not determined | 0.149 |
| ORF1–338 L93N/L199N: none (19) | ||||
| ORF11–338 E89A/L90A/M91A: none (42) | ||||
| ORF11–338 E92A/L93A/K94A: none (42) |
The N-terminal 53 residues of ORF1 are intrinsically disordered
ORF1 residues 1–53 are predicted to be disordered using both long-range and short-range disorder prediction algorithms (IUPRED and SPOT-Disorder-Single (44,45)), which is in agreement with the reported random coil signal of ORF11–51 in circular dichroism experiments (19). To directly interrogate the structure of the ORF1 N-terminal region with residue-by-residue resolution, we used solution NMR spectroscopy on samples of ORF11–53. The backbone amide resonances of ORF11–53 are situated in a narrow chemical shift range of 8.6–7.9 ppm in 1H in the 15N–1H heteronuclear single quantum coherence (HSQC) correlation spectrum, which is characteristic of disordered proteins (Fig. 2 B). The chemical shifts of Cα and Cβ are sensitive to partial secondary structure formation. Using two measures for predicting secondary structure from the observed chemical shift, we find the N-terminal region is predominantly disordered but with slight extended/β-strand character at the extreme N-terminus, peaking around K21 (δ2D, ∼25%) and slightly positive values in secondary structure propensity consistent with some helical propensity (δ2D, ∼10%) between residues E41 and R49 (Figs. 2 C and S8; (39,40)).
Because formation of partial structure should slow local reorientational motions in parts of ORF1, we acquired 15N-spin relaxation data to examine the motion of ORF11–53 at each residue position. The relaxation parameters are not uniform across the entire protein sequence and are significantly elevated after residue Q32 in two pH conditions. The average 15N-longitudinal relaxation rate constants, R2, and heteronuclear NOE ratio parameters at pH 6.0 for residues of region 1–31 (1.32 ± 0.12 s−1, 2.08 ± 0.33 s−1, and −0.40 ± 0.23) increase to average values of 1.74 ± 0.15 s−1, 3.34 ± 0.47 s−1, and −0.21 ± 0.14 for residues 32–53, with a similar trend at pH 7.25. Together, these data are consistent with some partial structures for residues 32–53. We also acquired 15N–1H HSQC correlation spectra at varying ORF11–53 concentrations. No chemical shift perturbations were observed with increasing protein concentrations from 20 to 700 μM, suggesting that the N-terminal region does not self-associate and exists as a monomer in solution (Fig. S9).
The N-terminus of ORF1 is necessary but not sufficient for phase separation and interacts with the coiled-coil domain
Although deletion of the N-terminal region severely impairs LLPS, ORF11–53 does not self-assemble in solution and is incapable of phase separation alone (Fig. S10). These observations imply that a component of the coiled-coil domain may be necessary to promote ORF1 condensation. Because the coiled-coil domain is also insufficient for phase separation alone, we assayed the ability of the individual domains to promote phase separation in trans by DIC microscopy (Fig. S11). Titrating higher concentrations of the N-terminus into the coiled-coil domain did not promote phase separation up to a 1:6 molar ratio (Fig. 3 A). However, titration of ORF11–53 into full-length ORF11–338 decreased phase separation in a concentration-dependent manner, demonstrating that residues 1–53 can compete with the full-length protein for binding (Figs. 3 B and S12).
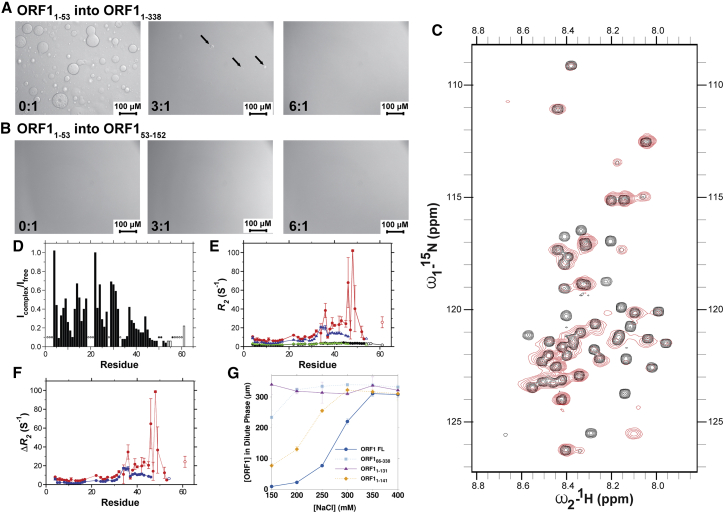
Figure 3.
The ORF1 N-terminal domain interacts with the C-terminal 22 residues of the coiled-coil domain. (A) ORF11–53 can disrupt phase separation of the full-length ORF11–338 protein in 300 mM NaCl, suggesting it competes for interaction sites found in ORF11–338 but cannot promote phase separation of the coiled-coil domain in trans, as shown in (B). Arrows in the 3:1 sample in (A) highlight the position of smaller droplets in the sample. This suggests that multivalency and interdomain interactions provided by the fusion of these domains is important for LLPS. All data were collected with 300 μM ORF11–338 or ORF153–152 at room temperature in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM DTT with increasing concentrations of ORF11–53. (C) Comparison of 15N–1H HSQC correlation spectra of 700 μM ORF11–53 alone (black) and 700 μM ORF11–53 mixed at an equimolar ratio with 700 μM ORF153–152 (red) at pH 6.0 (see detail below) shows line broadening of ORF11–53 in the presence of the coiled-coil domain with resonance attenuation consistent with binding of ORF153–152 around residues 47–53 of ORF11–53 and attenuations distributed across the entire sequence, shown in (D), consistent with binding at multiple sites. Open bars correspond to residues from the histidine tag. Elevated values for the 15N R2 relaxation rate constants (E) are observed in the presence of ORF153–152 at pH 7.25 (black) and pH 6.0 (red). Open circles correspond to residues from the histidine tag. (F) The difference in R2 in the presence and absence of ORF153–152, ΔR2, provides further evidence that residues 47–53 are perturbed by the addition of ORF153–152 because of slowed or conformational exchange because of binding to the structured, slow-moving coiled coil. (G) Quantification of the ORF1 protein concentration remaining in the dispersed phase after centrifugation demonstrates that ORF11–131 phase separates much less readily than full-length ORF11–338, whereas ORF11–141 LLPS is intermediate. N-terminal domain deletion ORF165–338 also significantly disrupts LLPS. NMR experiments were collected in 20 mM HEPES (pH 7.25), 200 mM NaCl, 1 mM DTT, 5% D2O or in 20 mM MES (pH 6.0), 200 mM NaCl, 1 mM DTT, and 5% D2O to optimize the signal/noise ratio. Error bars denote the standard deviation of three technical replicates. To see this figure in color, go online.
To address whether residue-specific interactions occur between the N-terminus and coiled-coil domain, we titrated ORF153–152 into 15N-isotopically enriched ORF11–53. The addition of ORF153–152 induces line broadening of the ORF11–53 spectrum compared with free ORF11–53. Peaks across the entire sequence, but especially in the transiently helical region after residue Q32, show significantly decreased intensities (Fig. 3, C and D). Furthermore, the 15N R2 relaxation rates are elevated around residues M35, S50, and N51 (Fig. 3, E and F). Because enhanced line widths and elevated transverse relaxation rates are consistent with either binding or conformational exchange, these data provide evidence that regions with lower signal intensity and higher relaxation rates correspond to residues contacting the coiled coil. It is important to note that the line broadening and enhanced 15N R2 are complementary, and enhanced 15N R2 is not observed for every residue for which line broadening is observed. It is possible that regions from residues 32 to 53 contain the primary binding site, whereas residues 1–31, which show reduced intensity but no large difference to 15N R2 relaxation, form transient contacts with the coiled-coil domain, leading to a chemical exchange-induced line broadening. We hypothesized that residues 32–53 form the primary interaction sites that mediate initial binding with ORF153–152; consequently, any changes within these sites should minimize the interaction between ORF11–53 and ORF153–152. The 15N–1H HSQC spectrum of a truncated ORF11–46 construct is superimposable on the HSQC spectrum of ORF11–53 in the unbound form, aside from the truncated region. At the same concentrations and conditions, significantly less line-width broadening is observed for ORF11–46 than for ORF11–52 in the presence of ORF153–152, indicating that the interactions with the coiled-coil domain are partially disrupted by the removal of residues 47–53 (Fig. S13). Furthermore, we observe similar interactions at residues 32–53 in the context of the ORF11–152 construct (Fig. S14). Based on these data, we hypothesized that ORF1 phase separation proceeds by transient, direct interactions between the N-terminal disordered region and the trimeric coiled-coil domain.
Truncation of the C-terminal 22 residues of the coiled-coil domain abolishes phase separation
To further investigate the nature of the intermolecular interactions between the N-terminal disordered region and the coiled-coil domain, we asked if a head-to-tail orientation could facilitate the intermolecular contacts we observed by NMR spectroscopy. This type of interaction would place the N-terminal disordered region close to the base of the coiled-coil domain. We therefore generated two constructs that truncated the C-terminal 12 or 22 residues, ORF11–141 and ORF11–131, respectively. Gel filtration showed that both of these constructs still trimerize in solution. Whereas ORF11–141 showed decreased phase separation compared with ORF11–152, ORF11–131 showed no phase separation by DIC in the conditions and concentrations assayed, irrespective of salt concentration (Figs. 2 A and S15; Table 1). These results were further quantified by measuring the amount of protein remaining in the dilute phase in different buffer conditions compared with the full-length ORF11–338 (Fig. 3 G).
To address whether the residue-specific interactions observed between ORF11–53 and the full-length coiled-coil domain ORF153–152 were disrupted by removing the C-terminal 22 residues, we collected the 15N–1H HSQC spectrum of 15N-isotopically enriched ORF11–53 and added an equimolar ratio of ORF153–131. In these conditions, we did not observe line-width broadening (Fig. S16) as we observed after the addition of ORF153–152. Thus, these data are consistent with a model in which interactions between residues 47–53 and 132–152 are required for ORF1 phase separation.
Mutations within both the N-terminus and coiled-coil domain modulate the properties of ORF1 phase separation
We next examined the contribution of specific residues or residue types to ORF1 phase separation. Previous mutagenesis and deletion studies showed that the basic residues at the extreme N-terminus are necessary for L1 retrotransposition in HeLa cells (14,19,42,43) and that a triple replacement of G2, K3, and K4 to alanines or deletion of these three residues inhibited retrotransposition to the same extent as the polymerase-dead mutant D702Y in ORF2 (19,42). Unlike the disordered, low-complexity sequences observed in several phase-separating proteins, such as fused in sarcoma and TDP-43, the ORF1 N-terminus is neither composed of repeat units nor highly enriched in a particular residue type (35,46). Instead, ORF1 demonstrates charge patterning in the N-terminus with basic and acidic patches or clusters throughout the sequence, akin to the RNA helicase Ddx4 (Fig. S17; (47,48)). Based on our observation that increasing the ionic strength of the buffer outcompetes the protein-protein interactions, leading to ORF1 phase separation, we hypothesized that weak, transient interactions at these positively and negatively charged blocks may contribute to ORF1 phase separation. Therefore, we investigated the effects of mutations within the N-terminal region that have been documented to either inhibit or enhance retrotransposition of the L1 element in the context of full-length ORF11–338. We generated double and quadruple mutants at positions K3, K4, R7, and K8 to ask whether changes in the charge patterning in the N-terminus affect ORF1 phase separation. Replacing lysine residues with glutamates in the K3E/K4E double mutant reduced the ability of ORF1 to phase separate in conditions that are favorable for the wild-type protein but did not completely abolish phase separation at lower salt concentrations (50–150 mM NaCl). In the quadruple mutants, removing the N-terminal basic patch (K3A/K4A/R7A/K8A) or introducing negative charges (K3E/K4E/R7E/K8E) also reduced ORF1 phase separation (Figs. 4 and S18–S24, changes in charge patterning reported using κ-values shown in Table 1 (41)).
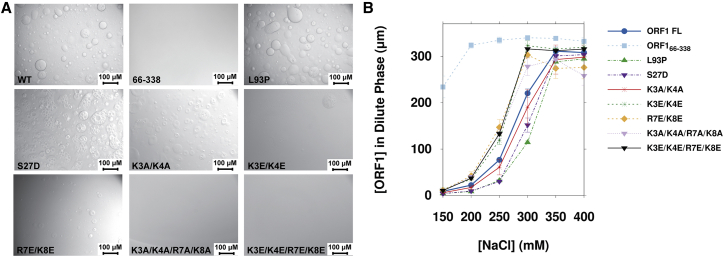
Figure 4.
Mutations in both the N-terminus and coiled-coil domain modulate phase separation of the full-length protein. (A) Altering the charge at the extreme N-terminus (K3E/K4E and K3E/K4E/R7E/K8E) impairs the ability of ORF1 to phase separate in 300 mM NaCl. Some mutations in the N-terminus and coiled-coil domain also appear to enhance the quantity and size of phase-separated droplets, as demonstrated by mutations S27D and L93P. All data were collected with 300 μM ORF1 at room temperature in 20 mM Tris (pH 8.0), 300 mM NaCl, and 1 mM DTT. (B) Quantification of ORF1 protein concentration in the dilute phase shows that mutations that introduce a positive charge at the extreme N-terminus significantly impair phase separation in comparison to the wild-type protein, whereas the S27D and L93P mutations enhance phase separation. Error bars denote the standard deviation of three technical replicates. To see this figure in color, go online.
In addition to the charge mutations, we examined the S27D phosphomimetic mutation because phosphorylation by proline-directed protein kinases increases L1 retrotransposition in cells (43). As observed by DIC microscopy, the S27D mutation increased the quantity and size of droplets compared with the wild-type protein, which is further substantiated by a reduced concentration of the S27D mutant protein in the dilute phase of the coexistence curve when compared with wild-type ORF1 (Figs. 4 and S25). In addition to increasing the propensity for phase separation, the S27D mutation also changes the morphology of the ORF1 phase-separated state as observed by DIC microscopy. Furthermore, we tested a naturally occurring L93P mutation in the stammer of the coiled-coil domain (49) and found enhanced phase separation in comparison with the wild-type protein, demonstrating that mutations within the coiled-coil domain also modulate the behavior of ORF1 phase separation (Figs. 4 and S26). Taken together, these data suggest that mutations in both the charged residues and coiled-coil domain modulate LLPS.
Discussion
The disordered N-terminal domain promotes liquid-liquid phase separation
Before our observations of ORF1 phase separation, no function had been proposed for the ORF1 N-terminal region, despite the fact that a charged N-terminus is highly conserved across species and that the extreme N-terminal residues are necessary for L1 retrotransposition (19,42). Here, we demonstrate that the full-length ORF1 protein forms liquid droplets in a salt-dependent manner and that the N-terminal region is disordered but required for this phase separation because it forms contacts with the coiled-coil domain. Differences in ORF1 phase separation demonstrated by both salt dependence and mutational studies altering charged residues in the N-terminal region suggest that electrostatic interactions are important for driving coalescence into the phase-separated state.
L1 is an ancient retroelement that has co-evolved with its eukaryotic hosts over vast stretches of evolutionary time. The ORF1 protein has adopted several functions and domains across evolutionary time in various species, including domain shuffling with functional host genes (50, 51, 52). Khazina and Weichenrieder (53) defined two main classes of ORF1 proteins: the type I family, found in plants, which contains an RRM and a CCHC zinc finger domain, and the type II family found primarily in vertebrates, composed of a coiled-coil domain, RRM, and conserved CTD. In addition, an array of variations within each group has been documented that do not fit into these classifications (53). For example, the Danio rerio ORF1 protein has gained an esterase domain that may serve a membrane-targeting function (54).
Human and mouse ORF1 belong to the type II family and have been functionally and structurally well characterized. The RRM and CTD act together to bind RNA, and the coiled-coil domain functions to trimerize the protein. Both are required for biological activity, namely to support the retrotransposition life cycle of the L1 element. The function of the largely disordered N-terminal region (residues 1–53) has remained enigmatic, although mutagenesis studies have clearly shown it to be essential for viability. We propose that the biological function of the N-terminal region is to enable multivalent self-interactions that lead to in vivo particle assembly and LLPS. Although the disordered N-terminal domain does not support LLPS alone, it acts in concert with the coiled-coil domain to enable LLPS of full-length ORF1. Although our in vitro LLPS conditions are unlikely to fully recapitulate LLPS in vivo, by biophysically characterizing a number of deletions as well as single and multiple point mutations, we demonstrated a strong correlation between LLPS and biological function (retrotransposition).
Sequence characteristics of the ORF1 N-terminal region
The N-terminal region shows the highest variability in length and sequence composition across species. However, a basic patch at the extreme N-terminus is highly conserved and is absolutely necessary for retrotransposition in human cells (19,42). Though less conserved than the basic patch, the charge around residues 47–53, which interacts with the base of the coiled-coil domain, is well conserved in vertebrates. Mutation of residues 44–46 (EEG) or 50–52 (SNY) to alanine reduces retrotransposition by ∼50% compared with wild-type ORF1 in cells in both cases (42). Aside from the deletion or mutation of the first three residues (GKK, which reduces retrotransposition to 4.6%), these two mutations in the disordered N-terminus show the greatest reduction on retrotransposition (44–46: 58.5% and 50–52: 53.8% (42)). This suggests that a reduction in the multivalent interactions seeded by the ORF1 N-terminus and coiled-coil domains may have a direct impact on the function of the L1 RNP particle.
The initial multivalency seeded by the interactions we observed using solution NMR spectroscopy between the base of the coiled-coil domains and the disordered N-terminus may be necessary to increase the local ORF1 concentration to nucleate phase separation. Once the concentration of ORF1 is sufficiently high, the intermolecular interactions mediated by the charged blocks within the ORF1 N-terminus then could establish a higher-ordered network that allows for demixing from the surrounding solution and provides environmental selectivity for LLPS. The coiled-coil domain also displays charged residues on the solvent-exposed surface because of the organization of the R-h-x-x-h-E repeat motifs and the constellation of charged residues in the x positions (Fig. 1 B; (19)). Although the arginine and glutamate at positions 2 and 6 of the repeat heptad are necessary for trimerization, the solvent-exposed charged residues may provide additional electrostatic interaction sites. The N-terminal domain may then form multiple electrostatic interactions with either the coiled-coil domain or possibly even N-terminal domains of neighboring ORF1 molecules in the phase-separated state. Hence, our work shows that the charged region 47–53 contributes significantly to interactions with the coiled-coil domain leading up to phase separation but that charged residues near the N-terminus also play a role in stabilizing the phase-separated form.
K3R and L93P are two naturally occurring polymorphisms among the 146 intact and active human L1 sequences in the L1Base2 database (49). Although these variants are competent for retrotransposition, removal of a single lysine at the N-terminus (K3) abolished retrotransposition in cell culture assays (19). The K3R polymorphism, in addition to maintaining the positive charge, may increase the interactions at the N-terminus because of the enhanced π-character introduced with the arginine mutation and has been shown to increase retrotransposition of the L1 element to 157% compared with the wild-type ORF1 protein (31,55). The L93P polymorphism within the stammer (residues 91–93) may further destabilize the coiled-coil domain and allow for increased flexibility at the N-terminus of the coiled-coil domain itself, which may help extend the disordered region when the beginning of the coiled coil is not fully populated. Khazina and Weichenrieder have demonstrated that an ORF11–103 construct is unstructured by circular dichroism spectroscopy and that the L93N mutation, which is predicted to rigidify the coiled coil, decreases retrotransposition (19). Further, mutating the residues in the stammer to alanines, which would extend and stabilize the coiled coil, abolishes retrotransposition to the same extent as deleting or mutating the N-terminal basic patch (89-ELM-91: 4.18%, 92-ELK-94: 0.69% (42)).
Alternating between the structured and unstructured states within the coiled-coil domain may provide a means for increasing the multivalency necessary for forming a phase-separated body. Elongating the disordered N-terminal region would allow for an extension of the N-terminus and increase any longer-range interactions with neighboring ORF1 molecules forming a higher-order mesh of the ORF1 protein. This would subsequently increase the local ORF1 concentration and allow for efficient biomolecular condensation.
Data availability
ORF11–53 resonance assignments are available in the Biological Magnetic Resonance Bank with BMRB: 50766.
Conclusion
Our data provide insights into the phase separation propensity and multivalent interactions between the N-terminus and coiled-coil domain and suggest a potential role for the structural malleability encoded within the stammer region of the coiled-coil domain. Further experiments will need to explore phase separation of the L1 RNP in cells and whether disrupting ORF1 phase separation directly impairs retrotransposition of the L1 element in vivo. Pharmacological inhibition of phase separation has emerged as a potential therapeutic approach (56), and modulation of ORF1 phase separation could provide an exciting novel strategy to interfere with L1 retrotransposition in aging and disease.
Author contributions
J.M.S. and G.J. conceived the study. J.C.N., M.T.N., N.L.F., J.M.S., and G.J. designed the experiments. J.C.N., G.Y.L., E.L.M., and M.T.N. performed the experiments. All authors contributed to data analysis and interpretation. J.C.N. wrote the manuscript with input from all authors.
Acknowledgments
J.M.S. is a cofounder of Transposon Therapeutics, serves as Chair of its Scientific Advisory Board, and consults for Astellas Innovation Management, Atropos Therapeutics, and Gilead Sciences. N.L.F. consults for and serves on the Scientific Advisory Board of Dewpoint Therapeutics.
This work was funded by National Institutes of Health R01 AG016694 and P01 AG051449 to J.M.S. and R01 NS116176 to N.L.F.
Editor: Samrat Mukhopadhyay.
Footnotes
Jocelyn Newton’s present address is The Hospital for Sick Children, Peter Gilgan Centre for Research and Learning, M5G 0A4 Toronto, Ontario, Canada.
Grace Li’s present address is Division of Hematology/Oncology, Boston Children’s Hospital, and Department of Pediatric Oncology, Dana-Farber Cancer Institute, Harvard Stem Cell Institute, Harvard Medical School, Boston, Massachusetts 02115.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.03.028.
Contributor Information
John M. Sedivy, Email: john_sedivy@brown.edu.
Gerwald Jogl, Email: gerwald_jogl@brown.edu.
Supporting material
References
- 1.de Koning A.P., Gu W., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levin H.L., Moran J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C.R., Burns K.H., Boeke J.D. Active transposition in genomes. Annu. Rev. Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns K.H., Boeke J.D. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancks D.C., Kazazian H.H., Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancks D.C., Kazazian H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erwin J.A., Marchetto M.C., Gage F.H. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat. Rev. Neurosci. 2014;15:497–506. doi: 10.1038/nrn3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns K.H. Transposable elements in cancer. Nat. Rev. Cancer. 2017;17:415–424. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- 9.De Cecco M., Criscione S.W., Sedivy J.M. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell. 2013;12:247–256. doi: 10.1111/acel.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Cecco M., Criscione S.W., Kreiling J.A. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013;5:867–883. doi: 10.18632/aging.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Cecco M., Ito T., Sedivy J.M. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566:73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon M., Van Meter M., Gorbunova V. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 2019;29:871–885.e5. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy B.K., Berger S.L., Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin S.L. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J. Biomed. Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mita P., Wudzinska A., Boeke J.D. LINE-1 protein localization and functional dynamics during the cell cycle. eLife. 2018;7:e30058. doi: 10.7554/eLife.30058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doucet A.J., Wilusz J.E., Moran J.V. A 3′ poly(A) tract is required for LINE-1 retrotransposition. Mol. Cell. 2015;60:728–741. doi: 10.1016/j.molcel.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin S.L., Bushman F.D. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khazina E., Truffault V., Weichenrieder O. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol. 2011;18:1006–1014. doi: 10.1038/nsmb.2097. [DOI] [PubMed] [Google Scholar]
- 19.Khazina E., Weichenrieder O. Human LINE-1 retrotransposition requires a metastable coiled coil and a positively charged N-terminus in L1ORF1p. eLife. 2018;7:e34960. doi: 10.7554/eLife.34960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathias S.L., Scott A.F., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 21.Moran J.V., Holmes S.E., Kazazian H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 22.Sokolowski M., Chynces M., Belancio V.P. Truncated ORF1 proteins can suppress LINE-1 retrotransposition in trans. Nucleic Acids Res. 2017;45:5294–5308. doi: 10.1093/nar/gkx211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodier J.L., Zhang L., Kazazian H.H., Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain S., Wheeler J.R., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor M.S., Altukhov I., LaCava J. Dissection of affinity captured LINE-1 macromolecular complexes. eLife. 2018;7:e30094. doi: 10.7554/eLife.30094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Youn J.Y., Dunham W.H., Gingras A.C. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Hubstenberger A., Courel M., Weil D. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Markmiller S., Soltanieh S., Yeo G.W. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beliakova-Bethell N., Beckham C., Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeynaems S., Alberti S., Fuxreiter M. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom A.R., Brangwynne C.P. The liquid nucleome - phase transitions in the nucleus at a glance. J. Cell Sci. 2019;132:jcs235093. doi: 10.1242/jcs.235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y.H., Forman-Kay J.D., Chan H.S. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry. 2018;57:2499–2508. doi: 10.1021/acs.biochem.8b00058. [DOI] [PubMed] [Google Scholar]
- 35.Burke K.A., Janke A.M., Fawzi N.L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang A., Conicella A.E., Fawzi N.L. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018;37:e97452. doi: 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson D.G., Young L., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 38.Horton R.M., Hunt H.D., Pease L.R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 39.Marsh J.A., Singh V.K., Forman-Kay J.D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sormanni P., Camilloni C., Vendruscolo M. The s2D method: simultaneous sequence-based prediction of the statistical populations of ordered and disordered regions in proteins. J. Mol. Biol. 2015;427:982–996. doi: 10.1016/j.jmb.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Holehouse A.S., Das R.K., Pappu R.V. CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adney E.M., Ochmann M.T., Boeke J.D. Comprehensive scanning mutagenesis of human retrotransposon LINE-1 identifies motifs essential for function. Genetics. 2019;213:1401–1414. doi: 10.1534/genetics.119.302601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook P.R., Jones C.E., Furano A.V. Phosphorylation of ORF1p is required for L1 retrotransposition. Proc. Natl. Acad. Sci. USA. 2015;112:4298–4303. doi: 10.1073/pnas.1416869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dosztányi Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018;27:331–340. doi: 10.1002/pro.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanson J., Paliwal K., Zhou Y. Accurate single-sequence prediction of protein intrinsic disorder by an ensemble of deep recurrent and convolutional architectures. J. Chem. Inf. Model. 2018;58:2369–2376. doi: 10.1021/acs.jcim.8b00636. [DOI] [PubMed] [Google Scholar]
- 46.Conicella A.E., Zerze G.H., Fawzi N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nott T.J., Petsalaki E., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brady J.P., Farber P.J., Kay L.E. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. USA. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penzkofer T., Jäger M., Zemojtel T. L1Base 2: more retrotransposition-active LINE-1s, more mammalian genomes. Nucleic Acids Res. 2017;45:D68–D73. doi: 10.1093/nar/gkw925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyshlyaev G., Voigt F., Novikova O. Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution. Proc. Natl. Acad. Sci. USA. 2013;110:20140–20145. doi: 10.1073/pnas.1310958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivancevic A.M., Kortschak R.D., Adelson D.L. LINEs between species: evolutionary dynamics of LINE-1 retrotransposons across the eukaryotic tree of life. Genome Biol. Evol. 2016;8:3301–3322. doi: 10.1093/gbe/evw243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saxton J.A., Martin S.L. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J. Mol. Biol. 1998;280:611–622. doi: 10.1006/jmbi.1998.1899. [DOI] [PubMed] [Google Scholar]
- 53.Khazina E., Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider A.M., Schmidt S., Weichenrieder O. Structure and properties of the esterase from non-LTR retrotransposons suggest a role for lipids in retrotransposition. Nucleic Acids Res. 2013;41:10563–10572. doi: 10.1093/nar/gkt786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vernon R.M., Chong P.A., Forman-Kay J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheeler R.J., Lee H.O., Hyman A.A. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv. 2019 doi: 10.1101/721001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ORF11–53 resonance assignments are available in the Biological Magnetic Resonance Bank with BMRB: 50766.