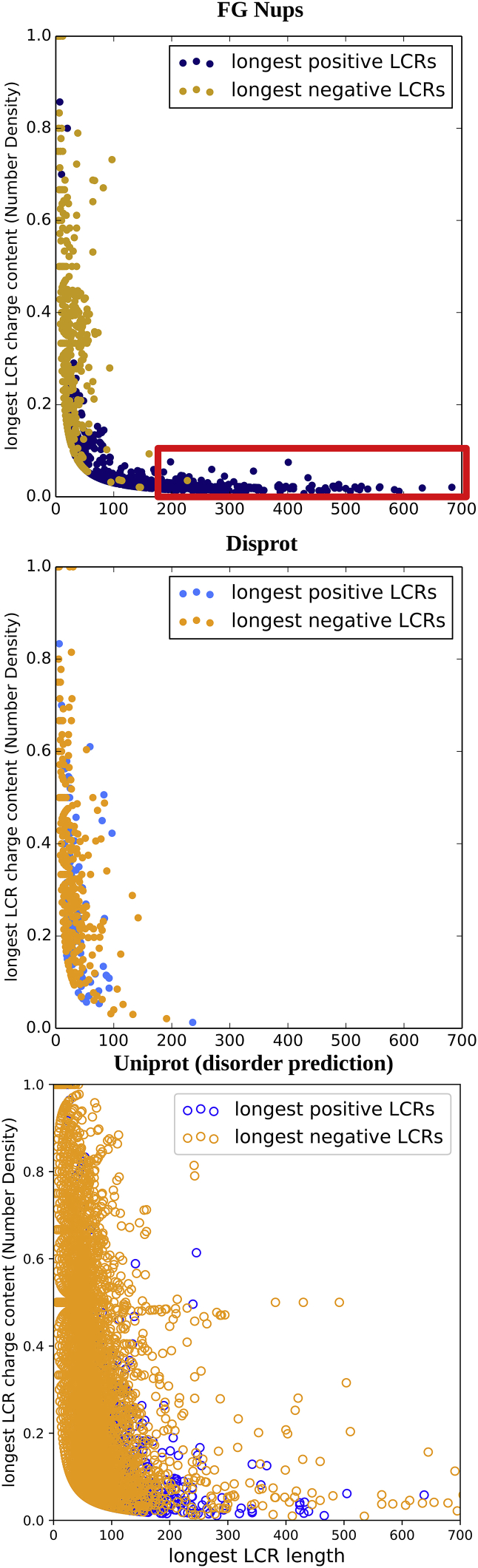
Figure 3.
Scatterplots showing charge content plotted against LCR length for each data set. Three data sets (FG Nups, DisProt, and UniProt (disorder prediction)) are compared with each other in terms of their length and charge content of their LCRs. FG Nups are removed from disorder prediction of UniProt for a more accurate comparison. Please see the Materials and methods for definition of each data set. Only the longest positive and negative LCRs from each sequence were plotted, and only if the length was greater than two amino acids. The data show a clear distinction between FG Nups and the other two data sets. Only FG Nups feature lpLCRs that are long but have a low charge density (boxed with red). It is important to note that this pattern does not exist in longest negative LCRs in FG Nups, DisProt, or UniProt (disorder prediction). Therefore, these data suggest that the specific pattern observed in lpLCRs is specific to FG Nups and an extremely small fraction of other proteins. The outliers comprise around 100 proteins, 40% of which are FG Nup ORFs. Another 20% are in p21-activated kinase family or of miscellaneous category, and the other 40% are hypothetical or predicted proteins with no known function. These data suggest that almost 0.01% of proteins in disorder prediction of UniProt are proteins that are not known FG Nups but feature lpLCRs that are larger than 200 amino acids. However, this percentage is significantly low, and our results suggest that lpLCRs are virtually exclusive to FG Nups. To see this figure in color, go online.