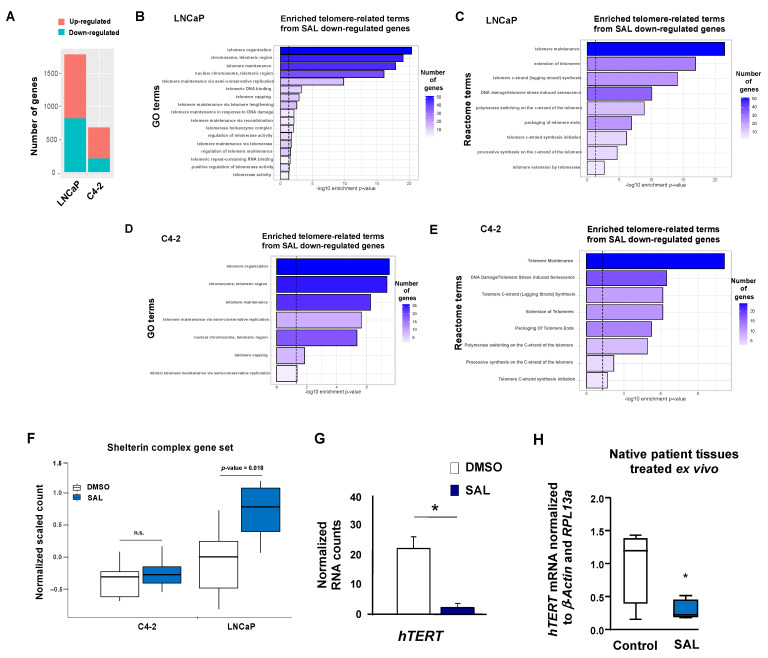
Figure 1.
Transcriptome analyses reveal a significant enrichment of telomeric Gene Ontology, Reactome terms, and shelterin complex subunits. The androgen-sensitive LNCaP or the castration-resistant C4-2 human PCa cells were treated with SAL (1 nM R1881) or solvent treated control (DMSO) 72 h prior to the isolation of RNA and performing RNA-seq. Transcriptome data were analyzed bioinformatically comparing SAL treated with solvent control-treated cells. (A) Number of differentially expressed genes (DEGs) including the number of upregulated and downregulated genes in both LNCaP and C4-2 cells. The telomere-related downregulated genes exhibit a stronger response to androgen treatment in LNCaP cells considering both the number of DEGs and the intensity of change (log2 fold). Indicated are SAL-mediated down-regulated genes significantly enriched in telomere-related GO and Reactome terms in LNCaP (B,C) and C4-2 (D,E) cells. (F) Gene set enrichment analysis (GSEA) of telomere-related genes, the subunits of the shelterin complex, upon SAL treatment compared with DMSO. Error-bars represent SEM. Wilcoxon paired test (gene-wise comparison, two-tailed) is used for p-value calculation. (G) mRNA expression of hTERT in LNCaP cells treated with the solvent control DMSO or SAL. Normalized RNA counts are indicated with the values for DMSO as the control. Adjusted p-value (* p < 0.05) was calculated using the DeSEQ2. (H) Native PCa tissue pieces obtained from patients with radical prostatectomy were treated ex vivo with R1881 at SAL as described previously (8). RT-qPCR was used to detect the regulation of hTERT expression by SAL. n = 4, one-tailed student t-test, * p-value: 0.028, n.s.: not significant.