Figure 40.
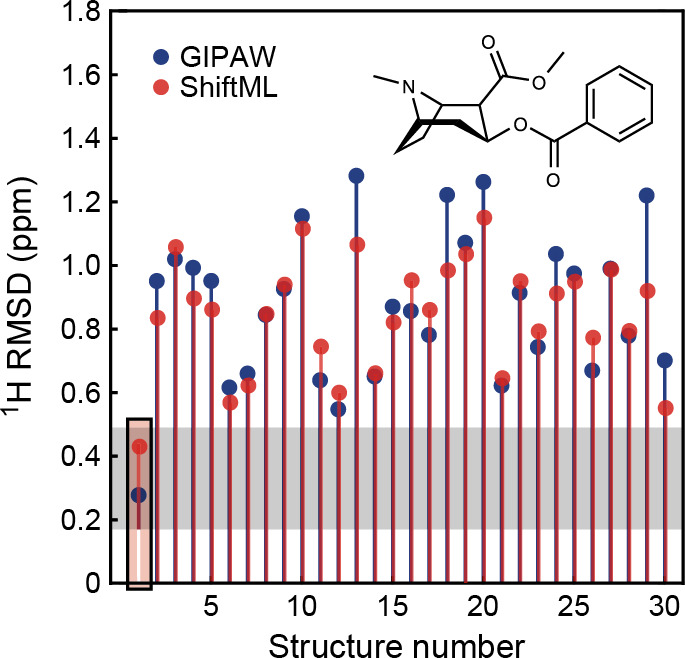
NMR chemical shift prediction. The figure shows a comparison between the 1H chemical shifts for a set of hypothetical polymorph structures of cocaine, obtained using a crystal structure prediction algorithm, and those of the most stable polymorph, determined experimentally. For each candidate structure an aggregate RMSD is shown between experimentally measured shifts and shifts calculated using either GIPAW-DFT (blue) or ShiftML (red). The gray zone represents the confidence intervals of the GIPAW-DFT 1H chemical shift RMSD. Candidates that have RMSDs within this range would be determined as correct crystal structures using a chemical shift-driven solid-state NMR crystallography protocol. Adapted from ref (386). Original figure published under the CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/).
