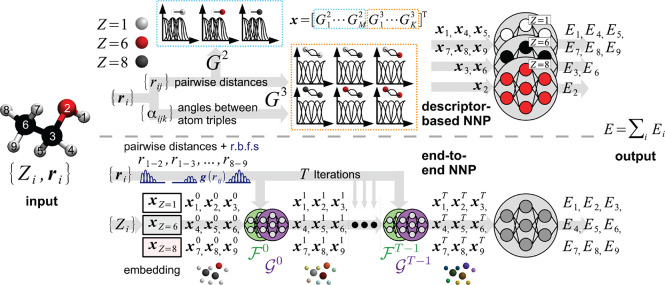
Figure 12.
Overview of descriptor-based (top) and end-to-end
(bottom) NNPs.
Both types of architecture take as input a set of N nuclear charges Zi and
Cartesian coordinates ri and
output atomic energy contributions Ei, which are summed to the total energy prediction E (here N = 9, an ethanol molecule is used
as example). In the descriptor-based variant, pairwise distances rij and angles αijk between triplets of atoms are calculated
from the Cartesian coordinates and used to compute hand-crafted two-body
(G2) and three-body (G3) atom-centered symmetry functions (ACSFs) (see eqs 22 and 23). For each atom i, the values of M different G2 and K different G3 ACSFs are collected in
a vector xi, which serves
as a fingerprint of the atomic environment and is used as input to
an NN predicting Ei.
Information about the nuclear charges is encoded by having separate
NNs and sets of ACSFs for all (combinations of) elements. In end-to-end
NNPs, Zi is used to initialize
the vector representation xi0 of each atom to an element-dependent
(learnable) embedding (atoms with the same Zi start from the same representation). Geometric
information is encoded by iteratively passing these descriptors (along
with pairwise distances rij expanded in radial basis functions g(rij)) in T steps
through NNs representing interaction functions 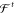 and
atom-wise refinements
and
atom-wise refinements 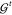 (see eq 25). The final
descriptors xi are used as input for
an additional NN predicting
the atomic energy contributions (typically, a single NN is shared
among all elements).
(see eq 25). The final
descriptors xi are used as input for
an additional NN predicting
the atomic energy contributions (typically, a single NN is shared
among all elements).