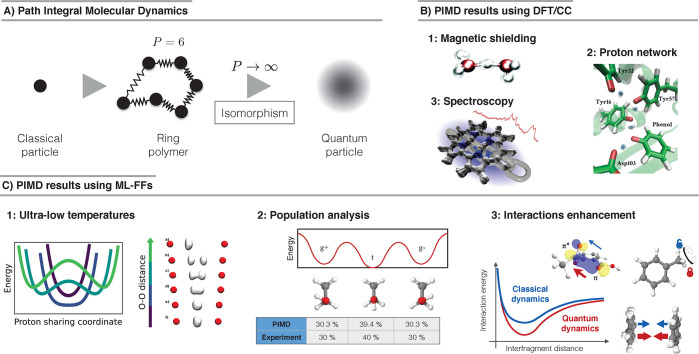
Figure 19.
(A) Schematic description of the path integral (ring polymer) molecular dynamics (PIMD) method, where quantum particles are approximated by a classical ring polymer with P beads. There is an exact isomorphism between these two systems when P → ∞, that is, their statistical properties become equivalent. (B) PIMD simulations using DFT or coupled cluster calculations. (1) Coupled cluster PIMD simulations of the Zundel model to compute 1H magnetic shielding tensor (adapted with permission from ref (287). Copyright 2015 published by the PCCP Owner Societies under CC BY-NC 3.0 https://creativecommons.org/licenses/by-nc/3.0/.). (2) Example of hydrogen-bond networks and their NQE implications on biological functions and enzyme catalysis (adapted with permission from ref (288). Copyright 2017 American Chemical Society.). (3) IR spectrum of the porphycen molecule computed from PIMD simulations (adapted with permission from ref (289). Copyright 2019 American Chemical Society.). (C) PIMD simulations using ML-FFs trained on DFT or coupled cluster data. (1) Ultralow temperature dynamics of the Zundel model obtained from PIMD simulations (adapted with permission from ref (290). Copyright 2018 American Chemical Society.). (2) Comparison of the statistical sampling of different conformers of ethanol between experiment and simulations (adapted with permission from ref (69). Copyright 2018 Chmiela et al.). (3) Schematic description of the enhancement in intra- and intermolecular interactions due to NQEs (adapted with permission from ref (161). Copyright 2020 Sauceda et al.).