Figure 6.
Overview of the mathematical concepts that form the basis
of kernel
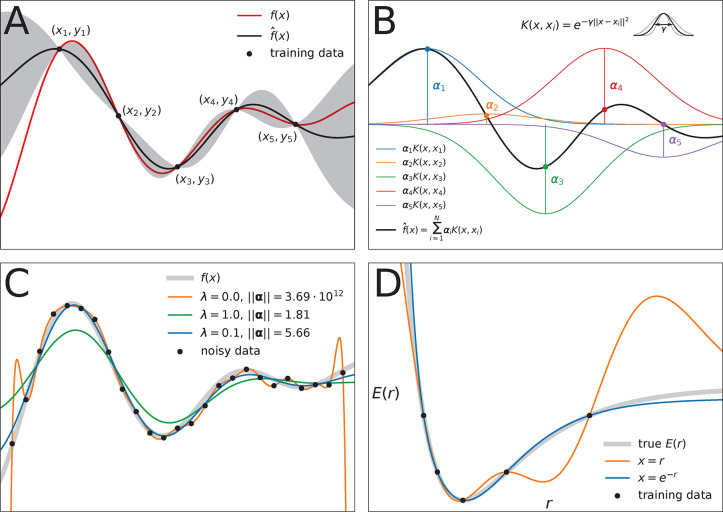
methods. (A) Gaussian process regression of a one-dimensional function f(x) (red line) from M = 5 data samples
(xi, yi). The black line 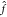 (x) depicts the mean (eq 8) of the conditional probability p(
(x) depicts the mean (eq 8) of the conditional probability p(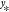 |y)
(see eq 7), whereas the
gray area depicts two standard
deviations from its mean (see eq 9). Note that predictions are most confident in regions where
training data is present. (B) Function
|y)
(see eq 7), whereas the
gray area depicts two standard
deviations from its mean (see eq 9). Note that predictions are most confident in regions where
training data is present. (B) Function 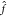 (x) can be expressed as
a linear combination of M kernel functions K(x, xi) weighted with regression coefficients αi (see eq 2). In this example, the Gaussian kernel (eq 4) is used (the hyperparameter γ
controls its width). (C) Influence of noise on prediction performance.
Here, the function f(x) (thick gray
line) is learned from M = 25 samples, however, each data point (xi, yi) contains observational noise (see eq 6). When the coefficients αi are determined without
regularization, i.e., no noise is assumed to be present, the model
function reproduces the training samples faithfully, but undulates
wildly between data points (orange line, λ = 0). The regularized
solution (blue line, λ = 0.1, see eq 10) is much smoother and stays closer to the
true function f(x), but individual
data points are not reproduced exactly. When the regularization is
too strong (green line, λ = 1.0), the model function becomes
unable to fit the data. Note how regularization shrinks the magnitude
of the coefficient vectors ∥α∥. (D)
For constructing force fields, it is necessary to encode molecular
structure with a representation x. The choice of this
structural descriptor may strongly influence model performance. Here,
the potential energy E of a diatomic molecule (thick
gray line) is learned from M = 5 data points by two kernel machines
using different structural representations (both models use a Gaussian
kernel). When the interatomic distance r is used
as descriptor (orange line, x = r), the predicted potential energy oscillates between data points,
leading to spurious minima and qualitatively wrong behavior for large r. A model using the descriptor x = e–r (blue line) predicts
a physically meaningful potential energy curve that is qualitatively
correct even when the model extrapolates.
(x) can be expressed as
a linear combination of M kernel functions K(x, xi) weighted with regression coefficients αi (see eq 2). In this example, the Gaussian kernel (eq 4) is used (the hyperparameter γ
controls its width). (C) Influence of noise on prediction performance.
Here, the function f(x) (thick gray
line) is learned from M = 25 samples, however, each data point (xi, yi) contains observational noise (see eq 6). When the coefficients αi are determined without
regularization, i.e., no noise is assumed to be present, the model
function reproduces the training samples faithfully, but undulates
wildly between data points (orange line, λ = 0). The regularized
solution (blue line, λ = 0.1, see eq 10) is much smoother and stays closer to the
true function f(x), but individual
data points are not reproduced exactly. When the regularization is
too strong (green line, λ = 1.0), the model function becomes
unable to fit the data. Note how regularization shrinks the magnitude
of the coefficient vectors ∥α∥. (D)
For constructing force fields, it is necessary to encode molecular
structure with a representation x. The choice of this
structural descriptor may strongly influence model performance. Here,
the potential energy E of a diatomic molecule (thick
gray line) is learned from M = 5 data points by two kernel machines
using different structural representations (both models use a Gaussian
kernel). When the interatomic distance r is used
as descriptor (orange line, x = r), the predicted potential energy oscillates between data points,
leading to spurious minima and qualitatively wrong behavior for large r. A model using the descriptor x = e–r (blue line) predicts
a physically meaningful potential energy curve that is qualitatively
correct even when the model extrapolates.