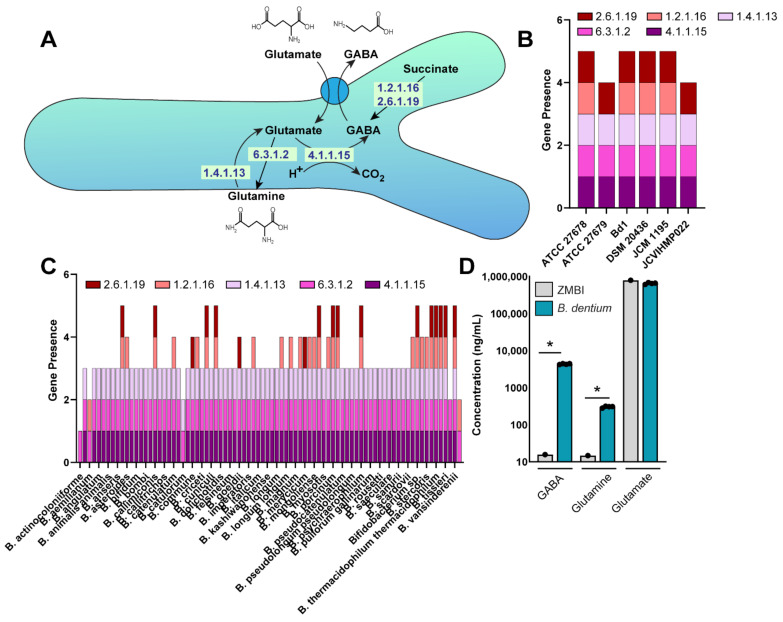
Figure 1.
(A). Gene pathways in Bifidobacterium dentium as identified via KEGG (https://www.genome.jp/kegg/kegg2.html) related to GABA, glutamate, and glutamine production. (B). Genome analysis of the JGI Integrated Microbial Genomes (IMG) database (http://img.jgi.doe.gov (accessed on 17 January 2021)) of six B. dentium genomes for the enzymes (Enzyme Commission, ECs) involved in glutamate, glutamine, and GABA production. Filled bars represent the presence of at least 1 gene copy of each enzyme. (C). Genome analysis of 83 Bifidobacteria genomes using the JGI Integrated Microbial Genomes (IMG) database. (D). Absolute concentrations of GABA, glutamate, and glutamine in uninoculated ZMB1 and cell-free bacterial conditioned ZMB1 from B. dentium after 18 h of growth. n= 4 biological replicates, two-way ANOVA, * p < 0.05.