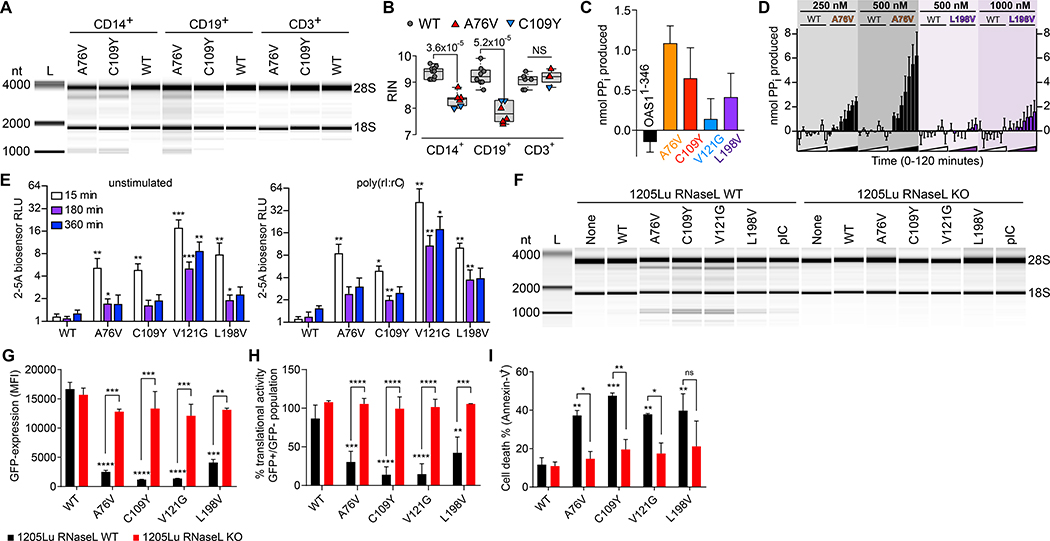
Fig. 4. OAS1-GOF 2–5A synthetase activity- and RNase L-dependent cellular RNA degradation, protein translational arrest, and apoptosis.
(A) RNA chip analysis from OAS1-WT, A76V, and C109Y monocytes (CD14+), B-cells (CD19+), and T-cells (CD3+) of total RNA degradation and (B) summary graph of RIN from 5 independent experiments (NS = non-significant). (C) In vitro activity for OAS1-WT and OAS1-MUT (A76V, C109Y, V121G, L198V) proteins in the absence of poly(rI:rC) dsRNA after 60 minutes (error bars, mean ± s.e.m). (D) Chromogenic analysis of reaction time courses for purified OAS1-WT (white bars), A76V (black bars), and L198V (purple bars) protein in the absence of poly(rI:rC) dsRNA for a range of protein concentrations (error bars, mean ± s.e.m). (E) 2–5A biosensor analysis of unstimulated and poly(rI:rC) dsRNA-stimulated HEK293T cells transiently transfected with OAS1-WT and OAS1-MUT (error bars, mean ± s.d.). (F) RNA chip analysis of total RNA isolated from 1205Lu melanoma RNase L wild-type (WT) and RNase L knockout (KO) cell lines untransfected (None) and transiently transfected with OAS1-WT, OAS1-MUT, or poly(rI:rC) (pIC). (G) Expression levels of OAS1-WT or OAS1-MUT in transfected 1205Lu melanoma wild-type (WT) and RNase L knockout (KO) cell lines determined by eGFP co-expression 48h after transfection (error bars, mean ± s.d.). (H and I) Translational activity (GFP-positive/GFP-negative) and frequency of dead (Annexin-V+) cells 48h after transfection (error bars, mean ± s.d., * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001).