Figure 1: Highlighted challenges and technologic advances in identification and validation of a direct MYCN targeting compound.
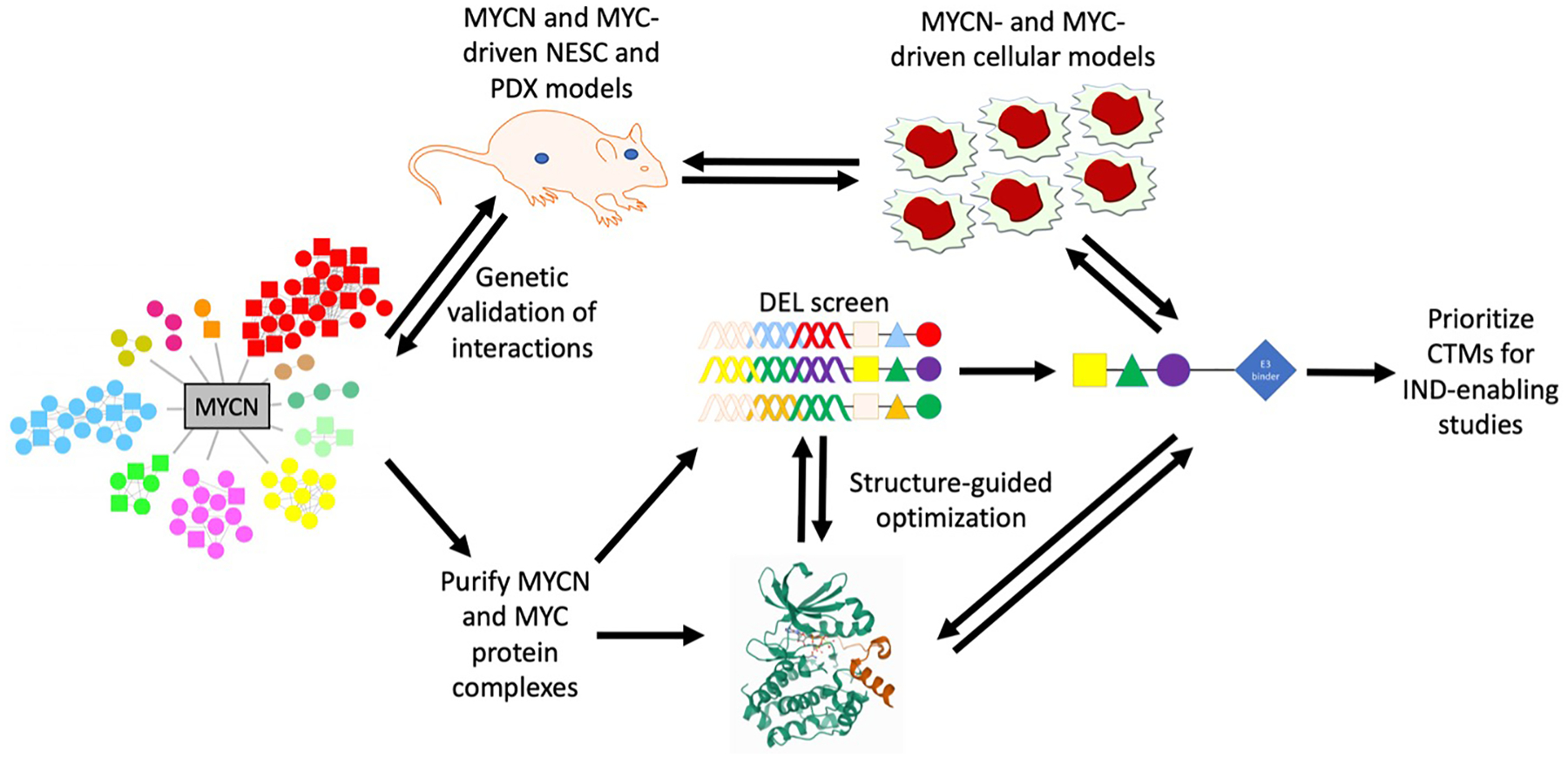
Biochemically-derived MYCN interactomes have recently revealed a large number of potential complexes to target and are contrasted to MYC interactomes (not shown). Which complexes are essential to tumor maintenance can now be identified using genetic loss-of-function testing in appropriate in vivo models (e.g. neural epithelial stem [NES] cell and patient-derived xenograft [PDX] models). Once oncogenic complexes are identified, these can be purified and used in structural studies, aided by advancements in cryo-EM technology. Challenges to the identification of small molecule inhibitors can be addressed by using a bind and degrade approach. DEL screens can facilitate the identification of binders unique to MYCN complexes as compared to MYC complexes. These compounds can then be linked to E3 ligase ligands to create proteolysis-targeting chimeras (PROTACs), optimized using structure as a guide, and tested in cell line and mouse models to confirm degradation, ensure anti-tumor activity, and determine selectivity. Once active and selective PROTACs are identified, they can be prioritized for IND-enabling studies and eventual clinical testing. Interactome modified from http://pennlab.ca/research/#, protein structure from the PDB (https://www.rcsb.org/structure/5G1X), and drawing tools from motifolio.com.
