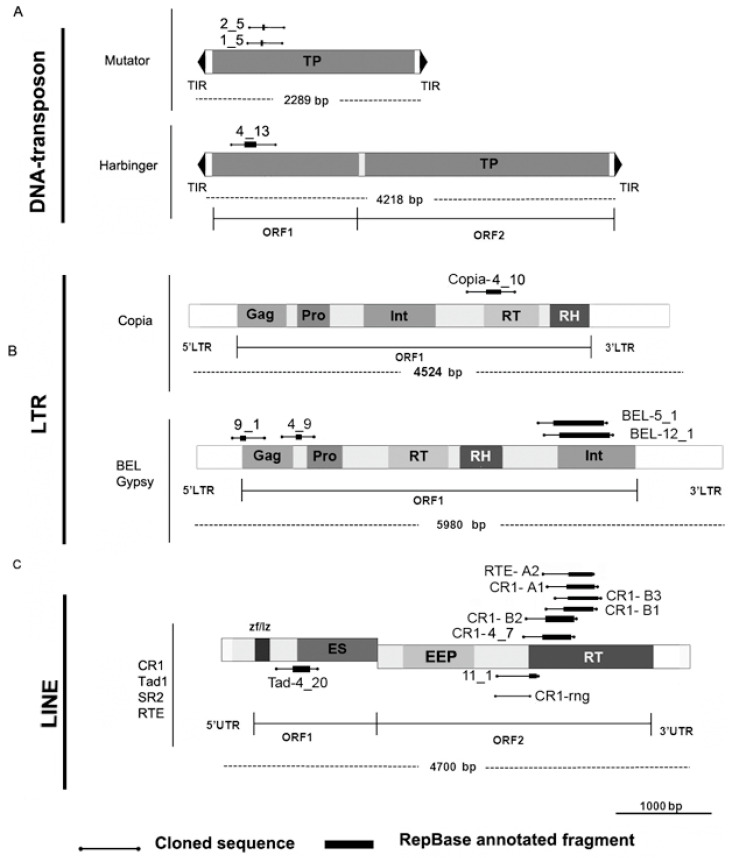
Figure 3.
Cloned fragments aligned to TE consensus sequences. (A–C) show transposon types, to which the cloned fragments belong. Element size, 5′- and 3′-untranslated regions (UTRs), terminal inverted repeats (TIRs), long terminal repeats (LTRs), and open reading frames (ORFs) are shown. Encoded predicted proteins: zf/lz domains—“zinc fingers” and “leucine zipper”; ES—esterase, EEP—exonuclease/endonuclease/phosphatase; RT—reverse transcriptase; RH—RNAseH; Int—integrase; Gag—DNA-binding protein; Pro—proteinase; TP—transposase. TE schemes were constructed according to [58], with modifications. The similarity of each RepBase hit is reported in Table S1.