On January 6, 2021, Guangdong CDC successfully isolated the 501Y.V2 variant of coronavirus disease 2019 (COVID-19) virus from a throat swab of a patient. The 501Y.V2 variant was first detected in South Africa, and the patient is a 55-year-old male airline pilot of South African nationality. He entered Guangzhou City, the capital of Guangdong Province, from Singapore on the evening of December 8, 2020 and was subject to a COVID-19 test. The result returned positive and he was transferred to the designated hospital for further isolation and treatment. On December 31, 2020, Guangdong CDC received the samples and began virus isolation and gene sequencing analysis. The analysis and subsequent verification have concluded that the virus genome belongs to lineage B.1.351, inferring classification as the 501Y.V2 variant. The patient is still receiving treatment.
On January 5, 2021, the sample and virus isolates were sequenced using Nanopore MK1C. Compared with the Wuhan reference strain (EPI_ISL_402119) (1-2), this strain displayed 23 nucleotide variation sites including the single nucleotide polymorphisms (SNPs) that defined the L-lineage European branch II.1 and belonged to the Pangolin lineage B.1.351 (3) (Figure 1). Furthermore, 10 amino acid mutation sites (D80A, L242del, A243del, L244del, R246I, K417N, E484K, N501Y, D614G, and A701V) were detected in the spike protein that corresponded to the features of the South African 501Y.V2 variant. The 501Y.V2 variant has 3 specific mutations, K417N, E484K, and N501Y, on the spike protein (4) and shares the N501Y and D614G mutations with the 501Y.V1 variant (also known as the B.1.1.7 lineage and variant of concern (VOC) 202012/01) recently detected in the United Kingdom. This is the second recent detection of a major international variant following the detection of the United Kingdom 501Y.V1 variant in a patient in Shanghai (NC20SCU2740-1) in December 2020 (5). The transmissibility and pathogenicity of these mutant variants urgently needs further study.
Figure 1.
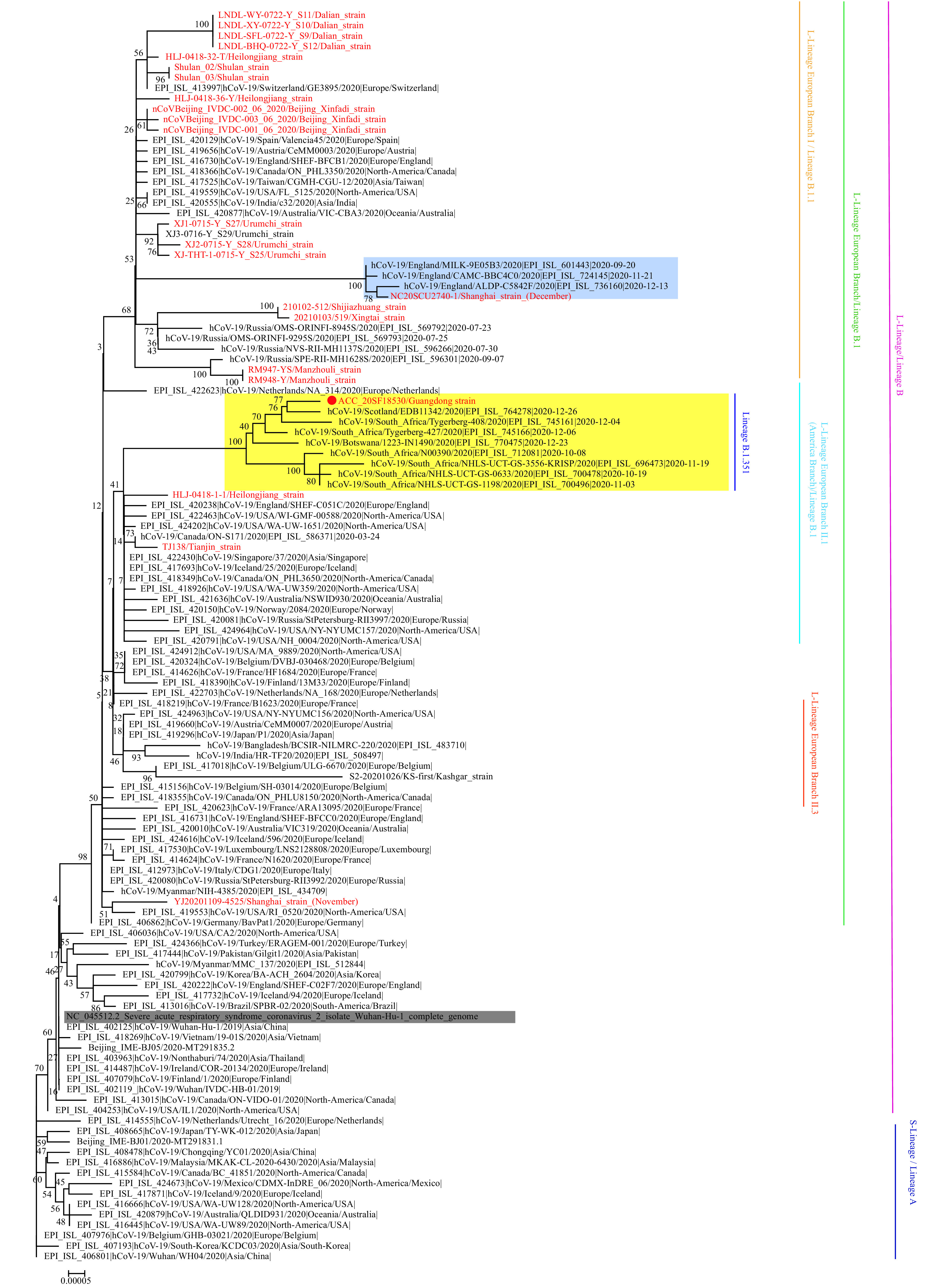
Phylogenetic tree based on the full-length genome sequences of the COVID-19 virus. Strains associated with specific outbreaks in China are marked using red letters. The South African 501Y.V2 variants are highlighted in yellow and the Guangdong imported 501Y.V2 variant is marked with a red dot. The UK VUI-202012/01 variants are highlighted in light blue. The Wuhan reference strain is shaded in gray. The S(A)- or L(B)-lineage and sublineages of the COVID-19 virus were marked and colored on the right.
Contributor Information
Chunliang Lei, Email: aaa.
Changwen Ke, Email: kecw1965@aliyun.com.
References
- 1.Tan WJ, Zhao X, Ma XJ, Wang WL, Niu PH, Xu WB, et al A novel coronavirus genome identified in a cluster of pneumonia cases — Wuhan, China 2019−2020. China CDC Wkly. 2020;2(4):61–2. doi: 10.46234/ccdcw2020.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–33. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al A dynamic nomenclature proposal for SARS-CoV-2 lineages to assistgenomic epidemiology. Nat Microbiol. 2020;5(11):1403–7. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GISIAD in the News. Novel variant 501Y.V2 with triple spike receptor binding site substitutions. https://www.gisaid.org/references/gisaid-in-the-news/novel-variant-combination-in-spike-receptor-binding-site/. [2021-01-07].
- 5.Chen HY, Huang XY, Zhao X, Song Y, Hao P, Jiang H, et al The first case of new variant COVID-19 originating in the United Kingdom detected in a returning student — Shanghai Municipality, China, December 14, 2020. China CDC Wkly. 2021;3(1):1–3. doi: 10.46234/ccdcw2020.270. [DOI] [PMC free article] [PubMed] [Google Scholar]


