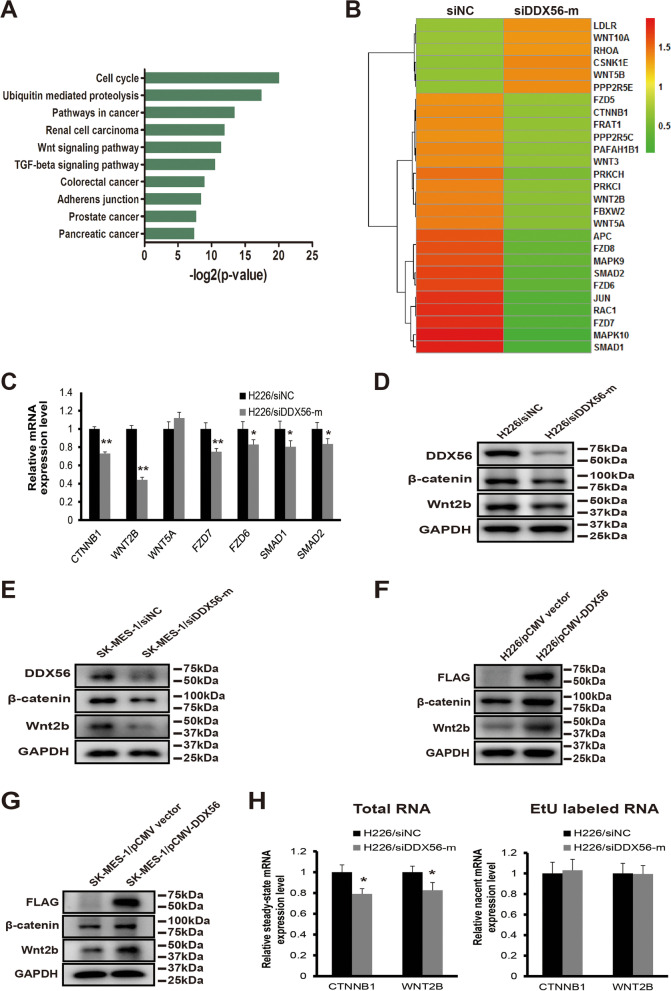
Fig. 4.
DDX56 post-transcriptionally regulates expression of the Wnt signaling pathway-related genes. a GO (KEGG) enrichment analysis of down-regulated mRNAs (> twofold) in a mRNA microarray experiment revealed that the KEGG terms “pathways in cancer” and “Wnt signaling pathway” were significantly enriched in down-regulated mRNAs in DDX56 siRNA- vs. control siRNA-transfected cells. b Heatmap showing WNT signaling-related genes differentially expressed (> twofold) between scramble siRNA (left) and DDX56 siRNA-treated (right) H226 cells in a mRNA microarray experiment. c qRT-PCR analysis of CTNNB1, WNT2B, WNT5A, FZD7, FZD6, SMAD1, and SMAD2 mRNA expression levels in DDX56-specific siRNA (siDDX56-mix) or negative control siRNA- (siNC) transfected H226 cells showed that DDX56 reduction significantly reduced mRNA expression of Wnt pathway-related genes CTNNB1, WNT2B, FZD7, FZD6, SMAD1, and SMAD2 except for WNT5A in H226 cells. d and e Western blot analysis of β-catenin, Wnt2b, and DDX56 protein expression level in H226 and SK-MES-1 cells transfected with DDX56-specific siRNAs or control siRNA, confirming reduced protein expression of β-catenin and Wnt2b following DDX56 reduction in H226 (d) and SK-MES-1 (e) cells. f and g Western blot analysis of the expression levels of β-catenin, Wnt2b and FLAG tag-fused DDX56 protein in H226 and SK-MES-1 cells transfected with pCMV-DDX56 or control pCMV vectors, confirming enhanced protein expression of β-catenin and Wnt2b following DDX56 overexpression in H226 (f) and SK-MES-1 (g) cells. GAPDH expression was used as a loading control. h Nascent transcript analysis showed no significant variation in the newly synthesized EtU-labeled RNA of CTNNB1 and WNT2B, whereas a significant decrease in the total (steady-state) RNA levels of CTNNB1 and WNT2B was observed after siRNA-mediated DDX56 knockdown in H226 cells. Gene expression levels were measured by qRT-PCR and standardized with GAPDH. Data are graphed as mean ± SD (n = 4). * P < 0.05