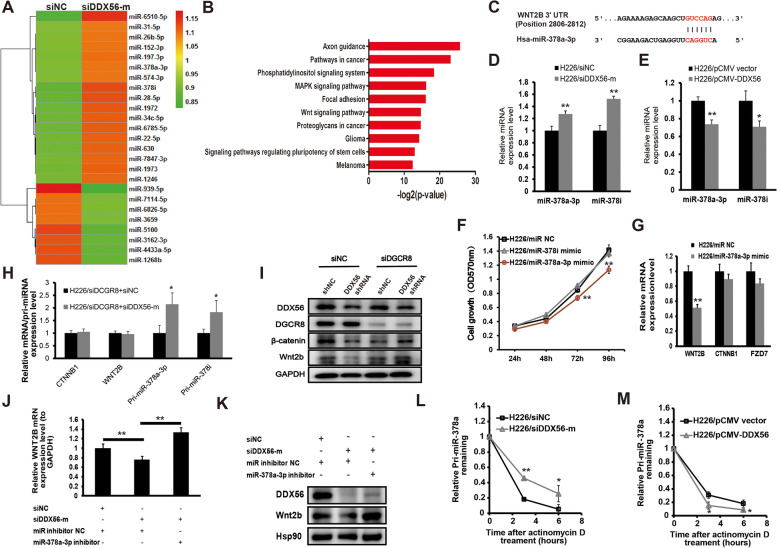
Fig. 5.
miR-378a-3p as a downstream effector of DDX56 involved in regulation of WNT2B expression in SqCLC cells. a Heatmap showing differentially-expressed miRNAs (≥ 1.2-fold) between scramble siRNA (left) and DDX56 siRNA-treated (right) H226 cells in a miRNA microarray experiment. b GO (KEGG) enrichment analysis of predicted targets of upregulated miRNAs (≥ 1.2-fold) in a miRNA microarray experiment revealed that the KEGG terms “pathways in cancer” and “Wnt signaling pathway” were also significantly enriched in predicted targets of up-regulated miRNAs in DDX56 siRNA- vs. control siRNA-transfected cells. c Predicted binding site of human miR-378a-3p to the 3’UTR of WNT2B by TargetScan. d and e qRT-PCR analysis of mature miRNA levels of miR-378a-3p and miR-378i in DDX56 silenced or overexpressed H226 cells compared to control cells. d H226 cells were transfected with DDX56-specific siRNA or control siRNA. e H226 cells were transfected with pCMV-DDX56 vector or pCMV empty vector. U6B was used for normalization of the mature miRNA expression data. (n = 3). f Analysis of growth of H226 cells transfected with miR-378a-3p mimic, miR-378i mimic or negative control miRNA mimic as determined by a crystal violet assay. (n = 4). g qRT-PCR analysis of WNT2B mRNA expression level in H226 cells transfected with miR-378a-3p mimic or control miRNA. GAPDH was used for normalization. (n = 3). h qRT-PCR analysis of mRNA expression of WNT2B and CTNNB1 and primary miRNA expression of miR-378a-3p and miR-378i in H226 cells co-transfected with DGCR8 siRNA and control siRNA or DGCR8 siRNA and DDX56 siRNA. GAPDH was used for normalization. i Representative Western blot analysis of Wnt2b and β-catenin protein expression levels in H226 cells co-transfected with control siRNA and control shRNA or control siRNA and DDX56 shRNA or DGCR8 siRNA and control shRNA or DGCR8 siRNA and DDX56 shRNA. GAPDH was used as a loading control. (n = 3). j qRT-PCR analysis of WNT2B mRNA expression level in H226 cells co-transfected with control miRNA and control siRNA or control miRNA and DDX56 siRNA or miR-378a-3p mimic and DDX56 siRNA. GAPDH was used for normalization. k Representative Western blot analysis of Wnt2b protein expression level in H226 cells co-transfected with control miRNA and control siRNA or control miRNA and DDX56 siRNA or miR-378a-3p mimic and DDX56 siRNA. Expression of a housekeeping gene, Hsp90 was used as a loading control. (n = 3). l and m Pri-miRNA stability assay carried out using H226 cells. l H226 cells were transfected with DDX56-specific siRNA (si-DDX56-mix) or control siRNA for 48 h, then treated with the transcription inhibitor actinomycin D. Primary miR378a were quantitated by qRT-PCR normalized to 18 s rRNA at 0, 3, and 6 h. (n = 3). m H226 cells were transfected with pCMV-DDX56 or empty pCMV vector for 48 h, then treated with the transcription inhibitor actinomycin D. Primary miR378a were quantitated by qRT-PCR, normalized to 18 s rRNA at 0, 3, and 6 h. (n = 3). Primary miR-378a expression level at 0 h was defined as 1. Each experiment was performed independently at least twice and results are presented as mean ± SD. Two-sided Student’s t-test was used to analyze the data. (*p < 0.05; **p < 0.01)