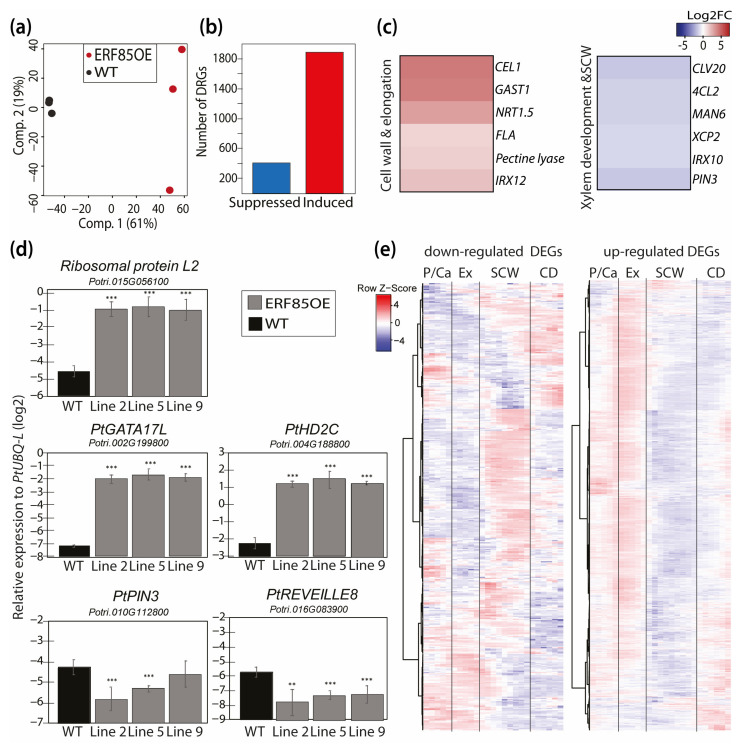
Figure 4.
Ectopic expression of PtERF85 stimulated the expression of genes linked to cell expansion and suppressed genes linked to SCW formation during xylem differentiation. (a) PCA of xylem transcriptome data obtained from three WT samples (WT, each sample represents a pool of three plants) and three ERF85OE lines (2,5,9). (b) Distribution of (DEGs in ERF85OE. DEGs were selected based on a |log2FC| > 1 and pAdj < 0.01 compared to WT. (c) Representative genes found among the DEGs in ERF85OE lines with known function of their homologs in Arabidopsis related to cell elongation, cell wall composition, and xylem development (Potri.015G127900 (CEL1); Potri.002G022600 (GAST1); Potri.001G171900 (Pectate lyase); Potri.008G064000 (IRX12); Potri.014G156600 (CLV20); Potri.010G112800 (PIN3); Potri.005G256000 (XCP2)) and SCW composition (Potri.010G192300 (FLA); Potri.001G036900 (4CL2); Potri.016G138600 (MAN6); Potri.005G256000 (IRX10)), Potri.003G088800 (NRT1.5). (d) Validation of differential expression of up- and down-regulated DEGs using qPCR. Bars represent the mean expression level+- SD of target genes relative to the housekeeping gene PtUBQ-L (log2). Asterisks indicate statistically significant differences between WT and ERF85OE lines according to the Student’s t-test (with p-value < 0.01 (**), and p-value < 0.001 (***)). (e) Heatmap showing the expression profile of up- and down-regulated DEGs during stem development in hybrid aspen. Data were obtained from the AspWood database [3]. Genes were scaled so that red represents the highest expression level for a given gene, and blue the lowest, across all developmental zones. P/Ca = phloem/cambium, Ex = expanding xylem, SCW = secondary cell wall formation, CD = cell death.