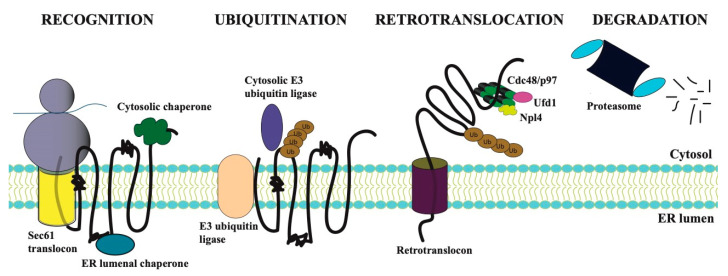
Figure 1.
The endoplasmic reticulum-associated degradation (ERAD) pathway consists of four steps. The degradation pathway for a generic integral membrane protein in the ER is shown, and misfolded regions in the lumen, membrane, and cytosol are represented within the polypeptide chain as condensed regions. Recognition: Integral membrane proteins enter the ER concomitant with protein translation on ER-associated ribosomes and with the assistance of the Sec61 translocon. A misfold region in a nascent polypeptide is recognized by cytosolic or lumenal chaperones. Ubiquitination: The ubiquitination machinery—and more specifically an E3 ubiquitin ligases, often along with an E2 ubiquitin conjugating enzyme (not shown)—is next recruited to the misfolded protein, which is then conjugated with a polyubiquitin chain. The chain most commonly contains Lys-48 isopeptide linkages, and a minimum of four ubiquitins is required for proteasome-dependent degradation [14]. Retrotranslocation: The ubiquitinated protein is retrotranslocated through an ER-integrated retrotranslocon. Retrotranslocation requires ATP-dependent extraction mediated by the Cdc48 (in yeast) or p97 (in mammals) complex. The Cdc48/p97 complex also consists of two associated factors, Npl4 and Ufd1, which aid in ubiquitinated substrate capture. Degradation: During and/or after retrotranslocation, the misfolded ubiquitination substrate is degraded by the 26S proteasome into short peptide fragments.