Figure 3.
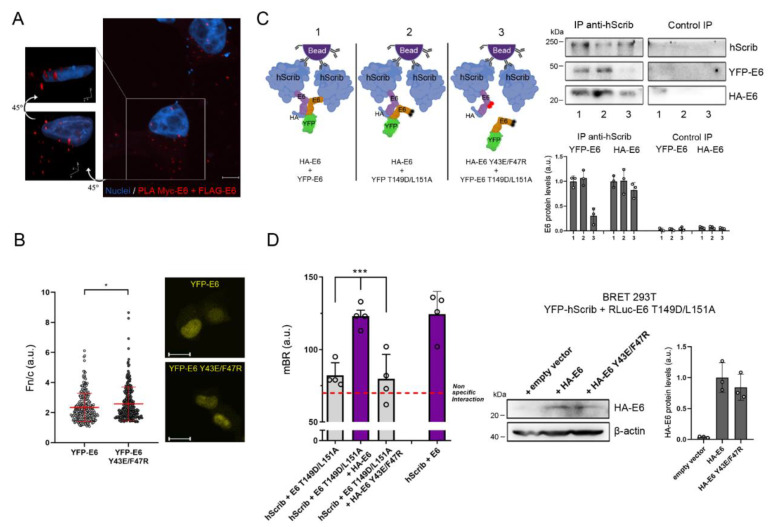
The dimeric form of E6 localizes in the cytosol and takes part in the E6–hScrib complex formation. (A) Representative confocal image of the Myc-FLAG Proximity Ligation Assay (PLA) detecting E6 homodimerization (red fluorescence) in H1299 cells co-expressing Myc-E6 and FLAG-E6. The image represents the maximum intensity projection of a Z-stack acquired with a 2000× magnification. Scale bar: 5 μm. Left panels show the three-dimensional reconstruction of the indicated cell and both images show the same cell, with a 45 degrees rotation around the horizontal axis between the two images. Nuclei were stained with DRAQ5. Experimental controls are shown in Figure S3A,B. (B) Quantitative nuclear/cytoplasmic fluorescence (Fn/c) analysis of H1299 living cells overexpressing YFP-E6 or YFP-E6 Y43E/F47R. Data are presented as scatter-dot-plots of nYFP-E6 = 247 cells and nYFP-E6 Y43E/F47R = 251 cells. * p < 0.05 determined with the Mann–Whitney U test. Right panels show the representative images used for quantification. Scale bars: 20 μm. (C) CoIP/Western blot analysis (right) of H1299 cells overexpressing YFP-tagged and HA-tagged E6 proteins showing the binding of tagged E6 to hScrib through immunoprecipitation of endogenous hScrib. Control IP represents the extent of non-specific binding of each protein in the experiment. Left diagram provides a schematic representation of the result obtained with the experiment, with red and black circles on E6 models indicating amino acid substitutions in the dimerization interface and PBM, respectively. Bar graphs (bottom) show relative densitometry quantifications of YFP-E6 and HA-E6 protein bands normalized to the respective precipitated hScrib. Data are Mean ± SD of 3 independent experiments. See Figure S3C for whole-cell extracts input controls. (D) Measurement of E6 T149D/L151A interaction with hScrib through BRET assays in HEK 293T cells in the absence or the presence of HA-E6 or HA-E6 Y43E/F47R (lower graph). The dashed line indicates the level of non-specific interaction of hScrib with an unrelated protein (UL44 of human cytomegalovirus). The interaction between hScrib and wild-type E6 was used as a positive control. Data are Mean ± SD of four independent experiments performed in triplicate. *** p < 0.001 determined with one-way ANOVA with Dunnett’s multiple-comparisons test. Right panel shows the Western blot analysis of HEK 293T cells subjected to BRET measurements, demonstrating the presence of the corresponding HA-tagged E6 protein in transfected cells. β-actin was used as a loading control. The bar graph shows the relative densitometry quantifications of HA-E6 protein bands normalized to β-actin. Data are Mean ± SD of 3 independent experiments. In (B–D), a.u.: arbitrary units.