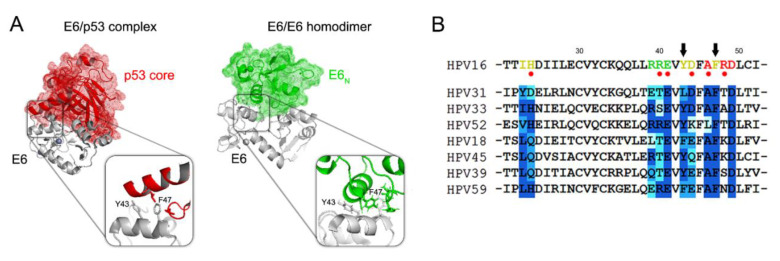
Figure 6.
Highly conserved hydrophobic residues of the α2 helix of HPV16 E6 mediate E6–p53 and E6–E6 interactions. (A) Ribbon/surface representations of the E6–p53 protein complex (left model; PDB code: 4xr8) and E6 homodimer (right model; PDB code: 2ljy). Panels show the contribution of Y43 and F47 residues for both interactions. In the model of E6 homodimer, the crystal structure of full-length HPV16 E6 (PDB code: 4giz) was superimposed on one of the two E6N monomers. Models created with PyMOL. (B) Alignment of the N-terminal amino acid residues (from 21 to 52 relative to HPV16 E6) of canonical high-risk E6 sequences. Residues of the α2 helix involved in E6 homodimerization (green), E6–p53 interaction (red) or both (yellow) are highlighted. Red circles indicate the α2 helix residues varying between HPV16 E6 variants in cervical cancer samples [57]. Black arrows indicate the critically conserved Y43 and F47 amino acids. Sequence alignment is shown according to Grantham’s distance coefficients, from dark blue (same or similar residues) to light blue (structurally dissimilar). Alignment generated with ClustalX.