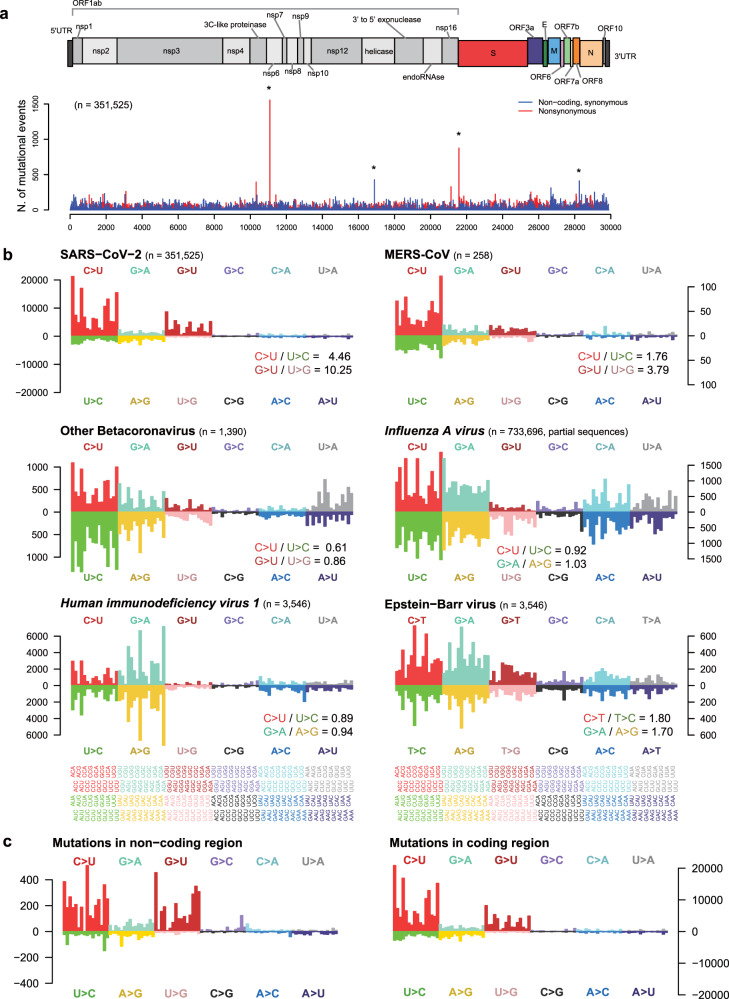
Fig. 1. Mutational signature of SARS-CoV-2.
a Distribution of the number of single-base substitutions along the viral genome. Each bar in the lower panel represents the counts for nonsynonymous substitutions (red) and synonymous substitutions in noncoding regions (blue). Except for a few recurrently mutated positions forming peaks, mutations are more or less uniformly distributed along the genome. The four highest peaks marked by asterisks are located in homopolymeric stretches. The highest peak at position 11,083 is caused by recurrent G→U and U→G substitutions in the context of U (5′-UUUUUUUGU-3′) (detailed in Supplementary Fig. 1). b Mutational signatures of SARS-CoV-2 and other viruses: MERS-CoV, other betacoronaviruses (including nonhuman hosted ones), Influenza A virus, HIV-1, and Epstein-Barr virus. For a given species (indicated in the panel title together with n = sample size), the panel shows the spectrum of observed substitution counts for 192 classes, a combination of changes of the major base (12 scenarios indicated with different colors) together with 4 types of the 5′ immediate upstream base and 4 types of the 3′ immediate downstream. Mutations of SARS-CoV-2 are particularly enriched in five sequence contexts (ACA, ACU, UCA, UCU, and GCU), and the mutational spectrum is asymmetric in terms of Watson-Crick base pairing and directional (i.e., the mutated and substituted bases are not balanced). Specifically, in SARS-CoV-2, C→U is much more frequent than U→C, and similarly, G→U is more frequent than U→G. MERS-CoV also exhibits an asymmetrical mutational spectrum similar to that of SARS-CoV-2. In contrast, Influenza A virus and HIV-1 show largely balanced patterns (C→U ≈ U→C and G→A ≈ A→G). Epstein-Barr virus, which is a DNA virus, shows asymmetry in its reversibility (C→T ≄ T→C) but exhibits symmetry in Watson-Crick base pairing (C→T ≈ G→A in a CpG context). c Comparison of mutational signatures in mutations of noncoding and coding regions.