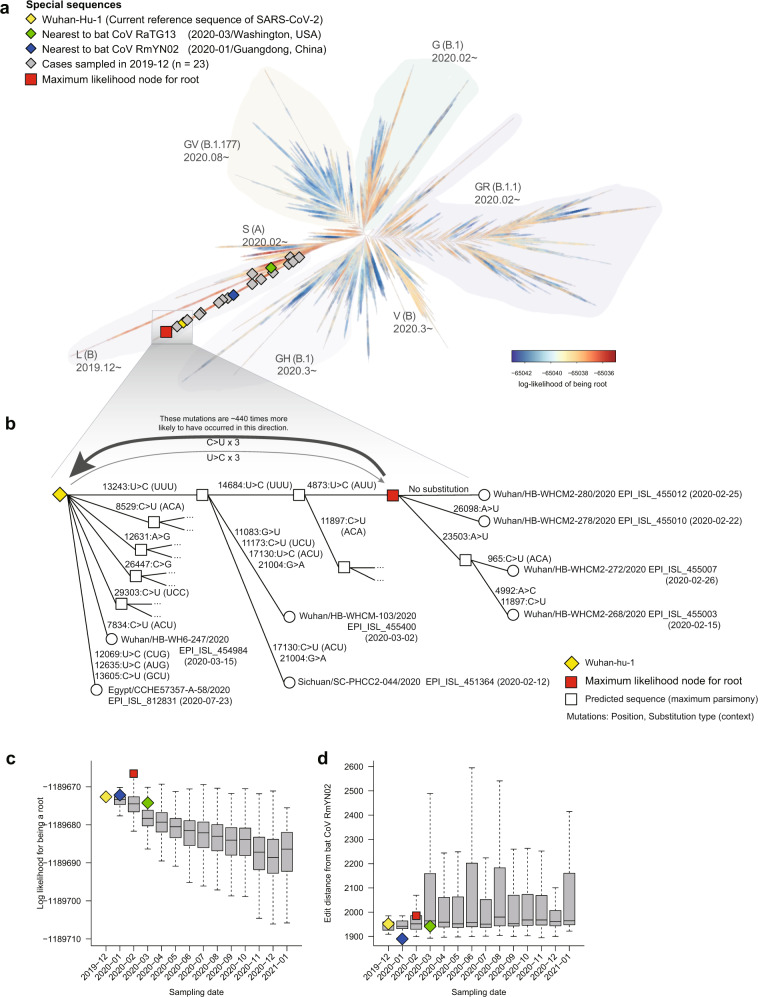
Fig. 2. Tracing the origin of SARS-CoV-2 using its mutational signature.
a Phylogenetic tree constructed from 351,525 full-length SARS-CoV-2 genome sequences deposited in the GISAID database. Well-known named clades are indicated with light shades of color and labeled according to widely used nomenclature75. Special sequences with prior knowledge: the Wuhan reference sequence (yellow), the sequence nearest to bat CoV RaTG13 (green) or RmYN02 (blue), and sequences sampled in December 2019 (gray) are marked with diamonds, and the presumed root (internal node with the maximum likelihood of being the root) is marked with a red square. The sequence data are constantly being updated on the GISAID website (https://www.gisaid.org/). b Detailed phylogenetic structure and associated substitutions between the presumed root (red square) and Wuhan-hu-1. c Boxplot showing the log-likelihood of being the root by collection time of the samples. Overall, likelihoods decrease steadily as collection time progresses. d Boxplot showing the edit distance from bat CoV RmYN02 by collection time of the samples. Overall, the edit distance does not significantly change over time.