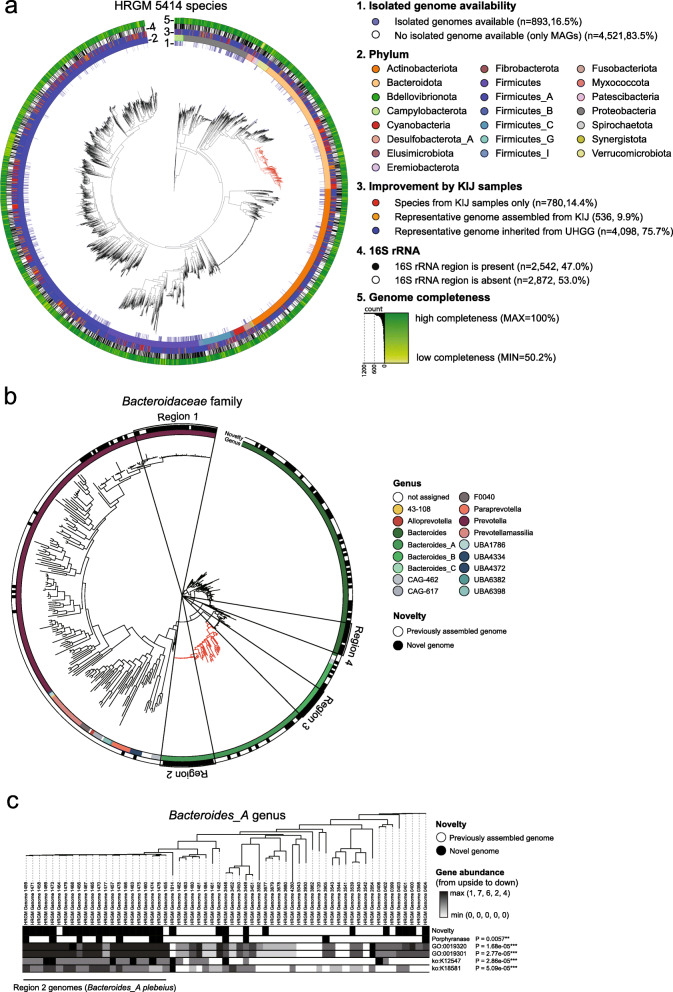
Fig. 2.
Phylogenetic trees of 5386 representative species of HRGM and species from KIJ samples. a Maximum-likelihood phylogenetic tree reconstructed from 120 bacterial marker genes (“Methods”). Representative genomes were annotated by their isolated genome availability (1st layer from the inside), phylum classification (2nd layer), whether they were from UHGG or assembled from KIJ samples (3rd layer), 16S rRNA sequence availability (4th layer), and genome completeness (the outermost layer). Red branches represent 410 species belonging to Bacteroidaceae family that are enriched in the representative species updated by including KIJ samples. b Bacteroidaceae family includes many new species from KIJ samples. The inner color strip indicates genus classification by GTDB-TK, and the outer color strip marks novel genomes from KIJ samples. Region 1 belongs to Prevotella genus and includes 30 species of Prevotella copri. Region 2 belongs to Bacteroides_A genus and includes 22 species of Bacteroides_A plebeius. Red branches indicate Bacteroides_A genus. Region 3 belongs to Bacteroides_B genus and 12 species of the region are annotated as Bacteroides_B vulgatus. Region 4 includes Bacteroides genus and all 13 species are classified as Bacteroides xylanisolvens by GTDB-TK. c Phylogenetic tree of 61 species of Bacteroides_A genus, the red branches in b. The uppermost color strip represents the novel species from KIJ samples. The second strip indicates the species harboring Porphyranase gene. Other gradient bars indicate the abundance of the gene with corresponding functional categories. P values indicate the statistical significance of enrichment of the function for the novel genomes compared with others in the Bacteroides_A genus. P values were calculated by the two-sided Fisher’s exact test (for the presence of Porphyranase) or two-sided Mann–Whitney U test (for abundance functional categories).