Abstract
Esophageal squamous cell carcinoma (ESCC) is the major subtype of esophageal cancer that is prevalent in Eastern Asia. Despite recent advances in therapy, the outcome of ESCC patients is still dismal. MicroRNAs (miRNAs) are non-coding RNAs which can negatively modulate gene expression at the post-transcriptional level. The involvement and roles of miRNAs have become one of the hot topics of cancer research in recent years. In ESCC, genetic variations within miRNA coding genes were found to have distinct epidemiological significance in different populations. Dysregulated expression of several miRNAs was reported to be associated with therapeutic response. Functionally, miRNAs can act either in an oncogenic or a tumor-suppressive manner during tumorigenesis of ESCC by interrupting signaling pathways associated with cell proliferation, metabolism, cancer stemness, and resistance to chemo- or radiotherapy. Moreover, miRNAs modulate metastasis of ESCC by targeting genes that regulate cytoskeleton dynamics, extracellular matrix remodeling, epithelial-mesenchymal transition, and tumor microenvironment. Most importantly, mounting evidence suggests that inhibiting oncogenic miRNAs or restoring the loss of tumor-suppressive miRNAs has therapeutic potential in the treatment of ESCC. Here, we review and discuss recent studies on the significance, biological functions, and therapeutic potential of miRNAs in tumorigenesis and metastasis of ESCC.
Keywords: MicroRNAs, Dysregulation, Tumorigenesis, Metastasis, Therapeutic potential, Esophageal squamous cell carcinoma
Core Tip: Esophageal squamous cell carcinoma (ESCC) is a deadly disease worldwide. Its poor prognosis is mainly due to the rapid tumor progression and high rate of invasion and metastasis. It is of great importance to understand the mechanisms underlying ESCC tumorigenesis and metastasis. Increasing studies confirmed the involvement of microRNAs (miRNAs) in cancer progression. Dysregulated miRNAs can serve as possible biomarkers for ESCC diagnosis or prognosis evaluation. Moreover, miRNAs function as small post-transcriptional regulators with notable therapeutic value. This review summarizes recent studies on the significance, biological functions, and clinical potential of miRNAs in tumorigenesis and metastasis of ESCC.
INTRODUCTION
Esophageal cancer is one of the most aggressive cancers worldwide. According to 2018 global cancer statistics based on 185 countries, esophageal cancer is the seventh most common cancer and the sixth in terms of mortality globally[1]. The two major subtypes of esophageal cancer, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC), differ in symptoms, geographic distribution, and etiology. ESCC is the most common subtype of esophageal cancer globally. It constitutes more than 80% of esophageal cancer cases worldwide[2-4]. In 2012, there were about 398000 new cases of ESCC, which was 7.6-fold higher than EAC cases[5]. Like many other cancer types, genetic alterations, uncontrollable proliferation, and escaping from cell death and immune-response are associated with the pathogenesis of ESCC. The high rates of local invasion and distant metastasis also contribute to the malignancy of ESCC. In fact, most patients have distant metastasis at initial diagnosis[6]. Delayed diagnosis and treatment lead to the poor prognosis. The 5-year survival rate of ESCC patients is only 20%-30%[7].
For decades, oncology studies mainly focused on dysfunction of protein-coding genes. It is only recently that the wide involvement of non-coding RNAs in cancer biology has been recognized. MicroRNAs (miRNAs) constitute one of the major families of non-coding transcripts. Typically, miRNAs are 20-24 ribonucleotides in length, and are transcribed from different genome locations, such as introns or junk DNA sequences between genes. MicroRNAs can bind to the 3’-untranslated region (UTR) of target mRNAs through imperfect base-pair complementation, and functionally promote target mRNA degradation or inhibit translation. The biogenesis of miRNAs is tightly controlled within normal cells, but mutations within miRNAs and dysregulated miRNA expression have been observed in many tumor types[8]. This highlights the possibility that miRNAs may be useful as diagnostic biomarkers. It has also been shown that miRNAs are involved in regulating multiple biological processes during cancer pathogenesis, which suggests that they may be exploited as therapeutic targets or tools[9]. In the last decades, increasing evidence supports that miRNAs have important roles in the pathophysiology of esophageal cancer[10,11]. In this review, we will summarize recent discoveries on the altered expression and functions of miRNAs in ESCC.
DYSREGULATION OF MICRORNAS IN ESCC
Genetic variations within miRNA sequence, as well as dysregulated level of miRNA, are frequently observed in multiple tumor types including ESCC[8]. Most of the genetic variations of miRNAs are due to the single nucleotide polymorphisms within miRNA coding genes (miR-SNPs)[12]. Studies in the last decades have demonstrated the epidemiological significance of miR-SNPs associated with susceptibility to ESCC in different populations. As a typical example, multiple studies reported that an SNP of miR-196a2, rs11614913 CC>TT, is associated with a reduced risk of ESCC in the Chinese Han population[13,14]. Another SNP of pro-miR-423, rs6505162 A>C, was found to be correlated with the incidence of ESCC in the Black population of South Africa. The same study also emphasized that this allele is strongly associated with inhaling smoke from burning biomass fuel, which is a known environmental risk factor for ESCC[15]. Functionally, polymorphisms of miRNAs can affect the expression of mature miRNA oligos by changing the secondary structure and stability of pre-miRNA molecules[12,16]. Taking polymorphism of miR-196a2 as an example, the rs11614913 TT genotype is associated with reduced miR-196a expression, which weakens its oncogenic function[13]. Besides, SNPs located in miRNA binding sites within the 3’UTR of cancer-related genes are also regarded as a critical issue of miRNA dysregulation. For instance, SNP rs6573A>C in the 3’UTR of RAS-related protein (RAP1A) was found to eliminate miR-196a binding, which consequently increased RAP1A expression and promoted ESCC development[17]. Another example is that CT or TT genotype of rs2866943, which is an SNP of protein tyrosine phosphatase receptor type T (PTPRT) 3’UTR, was reported to be associated with disrupted miR-218-regulated PTPRT expression[18].
Compared with genetic alteration of miRNA by SNPs, dysregulated miRNA expression patterns in ESCC have attracted much more attention in recent years. Yang et al[19] performed miRNA profiling in 113 pairs of ESCC tumor and matched non-tumor tissues, and found that 39 miRNAs were dysregulated in ESCC by at least 2-fold, including 28 downregulated and 11 upregulated miRNAs. Within the list of downregulated miRNAs, some (e.g., miR-133a and miR-133b) were also identified in other miRNA screening studies as possible tumor suppressors[20,21]. Dysregulation of several miRNAs was also reported to be associated with therapeutic response. For instance, miRNA profiling in ESCC tumor samples from patients receiving neoadjuvant radiochemotherapy showed that the expression of 12 miRNAs including 8 upregulated and 4 downregulated ones was altered in non-responders compared with responders[22]. In particular, high levels of miR-194 and miR-665, which were found in radiotherapy- or chemotherapy-resistant tumor samples, were correlated with a poor survival of ESCC patients, suggesting their potential as novel biomarkers for prognostic prediction[22]. Abnormal expression of miRNAs in ESCC may be due to many reasons. Genome instability is one of the underlying mechanisms. For example, Hu et al[23] performed global analysis of genome, mRNA, and miRNA status in 30 cases of ESCC, and demonstrated an association between bi-allelic loss in cancer genome and expression pattern change of 60 miRNAs. However, the authors also pointed out that bi-allelic loss in ESCC was infrequent, and that the relation between bi-allelic loss and miRNA dysregulation needs further validation[23]. Another piece of evidence is the location of some miRNA coding genes in the somatic copy-number alteration (SCNA) regions in cancer genome, such as miR-4448, miR-1224-3p, and miR-4707-5p in ESCC[24], which can lead to irregular mature miRNA expression. Moreover, several studies reported the correlation between dysregulated miRNA expression and abnormal epigenetic modulation in the promoter region. For example, promoter hypomethylation was found to enhance the expression of oncogenic miR-10-3p during ESCC development[25]. On the other hand, hypermethylation of CpG sites in promoter region was reported to downregulate the expression of tumor-suppressive miRNAs such as miR-375[26], miR-126[27], and miR-218[28,29] in ESCC. In addition, further studies demonstrated that miRNA expression is regulated by multiple upstream epigenetic factors, which is usually defined as “epigenetic-miRNA feedback loop”[30]. Taking miR-126 and miR-218 as examples, hypermethylation of their promoters is mediated by members of the DNA methyltransferase (DNMT) family, especially DNMT1 and DNMT3B[27,29]. Besides, epigenetic chromatin modifications also contribute to regulation of miRNA expression pattern. Koumangoye et al[31] reported that trimethylation of histone H3 lysine 27 (H3K27me3), which is a well-known heterochromatic modification, negatively regulated the expression of miR-31 in ESCC cells. In that study, the authors found that the SOX4/EZH2/HDAC3 co-repressor complex mediates hypermethylation of H3K27 around miR-31 promoter[31]. However, up to now, little is known about the regulatory function of other types of histone modifications on miRNA expression in ESCC. Further studies are needed to reveal the possible contribution of different histone modifications in modulating miRNA expression pattern in ESCC.
ROLES OF MICRORNAS IN TUMORIGENESIS OF ESCC
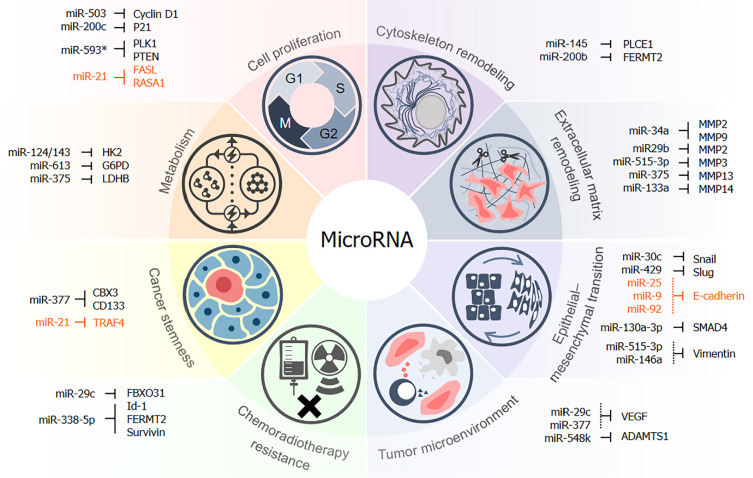
Beyond merely identifying alterations in miRNA expression patterns, researchers have become more focused on studying the functional significance of aberrantly expressed miRNAs. MiRNAs have been found to be involved in regulation of multiple biological processes during ESCC development (Figure 1). According to the distinct roles, miRNAs can be broadly described as tumor-suppressive and oncogenic miRNAs. Generally speaking, tumor-suppressive miRNAs functionally impede tumor malignancy, and are frequently repressed in expression level in tumor cells. For example, miR-503 was reported to negatively regulate cyclin D1 and induce cell cycle arrest in G1/S phase in ESCC[32]. Similar cell proliferation-repressive miRNAs include miR-200c and miR-593*, which interfere with mitosis by targeting P21 and target polo-like kinase 1 (PLK1), respectively[33,34]. Some miRNAs also suppress development of ESCC by regulating metabolic processes, especially glucose metabolism. For instance, miR-375 exerts a metabolism-repressive role by directly inhibiting lactate dehydrogenase B, which is a key enzyme in post-glycolysis process and catalyzes pyruvate-lactate interconversion[35]. Similarly, rate-limiting enzymes in glycolysis, such as glucose-6-phosphate dehydrogenase (G6PD) and hexokinase 2 (HK2), were reported to be negatively regulated by miR-613[36] and miR-125/143 cluster in ESCC[37]. Besides, miRNAs can also exert tumor-suppressive function by inhibiting cancer stemness. For example, Li et al[38] found that miR-377 was frequently downregulated in ESCC tumor, and that ectopic miR-377 expression could inhibit sphere formation and tumorigenic potential of ESCC cells by directly inhibiting CD133, which is a well-known stemness biomarker in cancer. In addition, it was reported that miR-377 can target chromobox protein homolog 3 (CBX3) and contributes to maintenance of stem cell potential in ESCC[39], which further emphasized the importance of miR-377 as a critical tumor suppressor in ESCC.
Figure 1.
Multiple roles of miRNAs during development of esophageal squamous cell carcinoma. This figure summarizes the targets and regulatory functions of miRNAs in multiple biological processes during esophageal squamous cell carcinoma (ESCC) development. MicroRNAs inhibit or promote tumorigenesis and recurrence of ESCC by modulating cell proliferation, metabolism, cancer stemness, and resistance to chemo- or radiotherapy. They also regulate metastasis by targeting functional molecules involved in epithelial-mesenchymal transformation, remodeling of the cytoskeleton, extracellular matrix, and tumor microenvironment. Oncogenic miRNAs and target genes are marked in orange. PLCE1: Phospholipase C epsilon 1; VEGF: Vascular endothelial growth factor.
Moreover, tumor-suppressive miRNAs can also promote the effect of chemoradiotherapies. A previous study by our group reported the tumor-suppressive role of miR-29c in reducing chemoresistance[40]. Overexpressing miR-29c in fluorouracil (5-FU)-resistant ESCC cell sublines increased their response to 5-FU treatment, shown as decreased cell viability and increased cell death, while miR-29c antagonist had opposite effect and could desensitize parental cells to 5-FU treatment. F-box only protein 31 (FBXO31), which has oncogenic function in ESCC[41] and lung cancer[42], was found to be one of the direct target genes of miR-29c that mediates its tumor-suppressive function. Further investigation found signal transducer and activator of transcription 5A (STAT5A) to be a transcription factor that can negatively regulate miR-29c expression in 5-FU-resistant cells[40]. This study highlighted the key role of the STAT5A/miR-29c/FBXO31 axis as a modulator of ESCC chemoresistance. By utilizing the same 5-FU-resistant cell model, Han et al[43] proved that miR-338-5p directly targets Id-1 (Inhibitor of DNA Binding 1) in ESCC cells to reverse chemoresistance. Interestingly, miR-338-5p was also able to attenuate cisplatin resistance and enhance radiotherapy efficiency by targeting focal adhesion protein kindlin-2 (also known as FERMT2)[44] and survivin[45], respectively. These studies suggest that miR-338-5p may be an essential tumor inhibitor.
On the other hand, onco-miRNAs are often upregulated in expression and functionally promote cancer progression by inhibiting tumor-suppressive proteins. One of the most well-studied oncogenic miRNAs in ESCC is miR-21. Several studies have reported that this miRNA is upregulated in the tumor[46-48], as well as in the serum or plasma of patients with ESCC[49-51]. Functionally, miR-21 was reported to promote cell growth and inhibit apoptosis in ESCC by activating the ERK/mitogen-activated protein kinase (MAPK) signaling cascade[52], or targeting genes including Fas ligand (FASL)[53], programmed cell death 4 (PDCD4)[54], phosphatase and tensin homolog (PTEN)[55-57], and RAS p21 protein activator 1 (RASA1)[58]. Further study also found miR-21 to play a role in maintaining cancer cell stemness in ESCC by upregulating stem cell markers such as Oct4 and Nanog and targeting TNF receptor-associated factor 4 (TRAF4)[59]. Oncogenic effects of miR-21 were also reported in breast cancer[60] and colorectal cancer[61], indicating its potential value as a pan-cancer therapeutic target. Onco-miRNAs can also interrupt cancer-related inflammation. For instance, miR-31 was found to be overexpressed in an orthotopic ESCC rat model promoted by Zn-deficiency, and functional tests revealed that miR-31 knockout abolished ESCC development and altered the expression profile of inflammation genes[62]. This effect was mediated by tumor suppressors egl-9 family hypoxia inducible factor 3 (EGLN3) and membrane bound o-acyltransferase domain containing 2 (MBOAT2)[62]. Actually, both oncogenic and tumor-suppressive functions of miR-31 were found in different cancer types[63], but knockout of miR-31 failed to show obvious genomic and metabolic instability in the rat esophagus, indicating that it might be of great value as a potential therapeutic target for ESCC[62].
ROLES OF MICRORNAS IN METASTASIS OF ESCC
The high incidence of metastasis is a serious issue associated with poor prognosis of ESCC. In addition to investigating the involvement of protein-coding genes in modulating tumor metastasis, recent studies have provided much information on the regulatory roles of miRNAs (Figure 1, also summarized in Table 1). MiRNAs were reported to modulate motility of ESCC cells by regulating cytoskeleton dynamics. For instance, miR-145 was demonstrated to have a suppressive effect on metastasis of ESCC by targeting phospholipase C epsilon 1 (PLCE1)[64], which is a Ras protein associated effector that can functionally remodel actin cytoskeleton[65,66]. Another example is miR-200b, which was reported to target FERMT2[67]. Therefore, the miR-200b/FERMT2 regulatory axis can reduce cell invasiveness by modulating the structure of the cytoskeleton and blocking the formation of focal adhesion[67].
Table 1.
Metastasis-regulating miRNAs in esophageal squamous cell carcinoma
|
MicroRNA
|
Validated target(s)
|
Effect
|
Ref.
|
| miR-1 | Notch2 | Represses proliferation, migration, and invasion | [105] |
| miR-9 | E-cadherin | Promotes metastasis | [91] |
| miRNA-10b-3p | TSGA10 | Promotes cell growth and metastasis | [106] |
| miR-17/20a | TGFBR2 | Represses migration and invasion | [107] |
| miR-21 | TGFβ | Promotes TGFβ-induced EMT | [108] |
| PCD4 | Promotes cell growth and invasion | [54] | |
| PTEN | Promotes invasion | [57] | |
| miR-25 | E-cadherin | Promotes metastasis | [93] |
| miR-26a and miR-144 | COX2 | Repress proliferation and metastasis | [109] |
| miR-29b | MMP2 | Represses proliferation and invasion | [73] |
| miR-29c | VEGF | Represses angiogenesis and metastasis | [95] |
| miR-30c | SNAI1 | Represses proliferation, EMT and invasion | [86] |
| miR-34a | MMP2, MMP9, and FNDC3B | Represses migration and invasion | [71] |
| CD44 | Represses invasion and metastasis | [110] | |
| Yin Yang-1 | Represses migration and invasion | [72] | |
| miR-92a | E-cadherin | Promotes lymph node metastasis | [92] |
| miR-92b | ITGAV | Represses invasion and metastasis | [111] |
| miR-106b | PTEN | Promotes invasion and metastasis | [112] |
| SMAD7 | Represses EMT | [84] | |
| miR-128-3p | ZEB1 | Represses EMT and metastasis | [89] |
| miR-130a-5p | ZEB1 | Represses EMT and metastasis | [113] |
| miR-130-3p | SMAD4 | Represses EMT, migration, and invasion | [82] |
| miR-133a | MMP14, FSCN1 | Represses cell invasion | [76] |
| miR-145 | PLCE1 | Represses proliferation and metastasis | [64] |
| miR-146a | Vimentin | Represses tumor invasion | [85] |
| miR-150 | ZEB1 | Represses EMT and metastasis | [90] |
| miR-200b | Kindlin-2 | Represses focal adhesion formation and invasion | [67] |
| miR-218 | BMI1 | Represses proliferation and metastasis | [114] |
| miR-339-5p | TSPAN15 | Represses metastasis | [115] |
| miR-375 | SHOX2 | Represses invasion and metastasis | [116] |
| MMP13 | Represses migration and invasion | [75] | |
| miR-377 | CD133 and VEGF | Represses tumor initiation and progression | [38] |
| miR-424-5p | SMAD7 | Represses EMT, migration and invasion | [83] |
| miR-429 | Slug | Represses migration and invasion | [87] |
| miR-515-3p | MMP3 and Vimentin | Represses invasion and metastasis | [74] |
| miR-548k | ADAMTS1 | Promotes lymph node metastasis | [96] |
| miR-630 | Slug | Represses invasion and metastasis | [117] |
| miR-644a | PITX2 | Repressing aggressiveness and stem cell-like phenotype | [118] |
| miR-655 | TGFBR2 and ZEB1 | Represses EMT | [88] |
| miR-1290 | SCAI | Promotes proliferation and metastasis | [119] |
| miR-4324 | FAK | Represses EMT | [120] |
| miR-6775-3p | MAGE-A family proteins | Represses invasion and metastasis | [121] |
TGF-β: Transforming growth factor β; PTEN: Phosphatase and tensin homolog; PLCE1: Phospholipase C epsilon 1; VEGF: Vascular endothelial growth factor.
The invasiveness of cancer cells is largely dependent on remodeling of the extracellular matrix (ECM) by matrix metalloproteinases (MMPs)[68]. Members of the MMP family are endopeptidases that catalyze the degradation of ECM components. In ESCC, highly expressed MMPs are often associated with strong malignancy and poor prognosis[69,70]. MiRNAs can modulate ESCC cell invasiveness by targeting MMPs. Yang et al[71] reported that a p53-downstream miRNA, miR-34a, suppressed ESCC cell migration and invasion by directly targeting and suppressing MMP2 and MMP9. Interestingly, miR-34a was also found to inhibit the expression of an upstream transcription factor of MMP2 and MMP9, namely, Yin Yang 1 (YY1), in ESCC cells, so that MMP2 and MMP9 levels were negatively regulated in ESCC[72]. Besides, MMP2 was also reported to be regulated by miR-29b in ESCC[73]. Qi et al[73] showed that the miR-29b/MMP2 axis inhibited cell invasion in vitro, as well as tumor growth in an animal model, indicating its involvement in both tumor development and metastasis. Other members of the MMP protein family, such as MMP3, MMP13, and MMP14, were found to be regulated by miR-515-3p[74] , miR-375[75], and miR-133a[76], respectively.
Apart from gain of cell motility and remodeling ECM by MMPs, epithelial-mesenchymal transition (EMT) is another important process during pre-metastasis stage. Generally speaking, EMT describes the loss of epithelial cell phenotype to assume a mesenchymal cell phenotype. It is considered to be a transitional process with a spectrum of stages, such as losing cell polarity and cell-cell junction, enhancing migratory property and invasiveness, and acquiring resistance to anoikis[77-79]. The EMT process is controlled by many growth factors, but the dominant inducer is considered to be transforming growth factor β (TGF-β)[80]. TGF-β stimulates Smad and MAPK signaling, thereby inducing a series of genetic events during EMT. Activation of SNAIL, ZEB, and TWIST transcription factor families is the initiation step. These transcription factors silence the gene expression of tight junction molecules of epithelial cells such as E-cadherin, tight junction protein 1 (TJP1), and zonula occludens 1 (ZO-1), and translationally activate the expression of mesenchymal marker molecules such as N-cadherin and vimentin[81]. Up to now, several miRNAs have been found to regulate the EMT process by directly targeting these related elements during metastasis of ESCC. For example, miR-130a-3p was reported to suppress EMT in ESCC by targeting SMAD4, which is one of the downstream molecules of the TGFβ signaling pathway[82]. Similarly, SMAD7 can be regulated by miR-424-5p[83] and miR-106b[84]. The miR-515-3p/vimentin regulatory axis can reverse EMT and negatively regulate ESCC metastasis[74]. Another miRNA that targets vimentin, namely, miR-146a, was also reported to suppress ESCC invasion by disrupting fibronectin membrane assembly[85]. Two members of the SNAIL transcription factors family, Snail and Slug, were found to be regulated by miR-30c[86] and miR-429[87], respectively. The transcription factor ZEB1 is negatively regulated in ESCC by miR-655[88], miR-128-3p[89], and miR-150[90]. On the contrary, miR-25, miR-9, and miR-92 act as oncogenic miRNAs that can promote EMT and ESCC metastasis by targeting E-cadherin[91-93].
Metastasis is affected by biochemical and cellular components surrounding tumor cells, which constitute the tumor microenvironment (TME). MiRNAs are functionally involved in the interaction between ESCC cells and the TME. On the one hand, the expression pattern of miRNAs in ESCC cells can be modulated by the TME, especially under stressful conditions such as hypoxia[94]. On the other hand, miRNAs are also known to regulate the crosstalk between ESCC cells and surrounding cancer-associated cells. In particular, several studies have found that miRNAs modulate the expression or secretion of vascular endothelial growth factor (VEGF). For instance, one of the previous studies in our group reported that miR-29c could suppress VEGF expression in cancer-associated fibroblasts, and the expression of miR-29c was in turn regulated by insulin-like growth factor 2 (IGF2) secreted by ESCC cells, suggesting the existence of a IGF2/miR-29c/VEGF regulatory loop which can promote ESCC distant metastasis[95]. Another study performed by our group found that VEGF was also targeted and inhibited by miR-377 in ESCC cells, so that the miR-377/VEGF axis had a negative impact on ESCC metastasis by downregulating angiogenesis[38]. In addition, Zhang et al[96] demonstrated that miR-548k directly inhibited metalloproteinase ADAMTS1, which acted as a chaperone and blocked intrinsic VEGFC. In addition, high level of miR-548k in ESCC cells facilitated VEGFC secretion. As a consequence, the miR-548k/ADAMTS1/VEGFC axis promoted lymph node metastasis by activating VEGFR3 in lymphatic endothelial cells[96].
THERAPEUTIC POTENTIAL OF MICRORNAS IN ESCC
In view of increasing evidence proving that miRNAs have important regulatory roles in cancer, miRNAs are considered as a pool of ideal therapeutic targets for developing novel treatment strategies. To be specific, treatment with synthetic antagonists or inhibitors may specifically neutralize and block the endogenous activity of oncogenic miRNAs. This is proposed as “mRNA suppression therapy”. As for tumor-suppressive miRNAs, “miRNA replacement therapy”, which is defined as restoring the loss of tumor-suppressive miRNA and enhancing the post-transcriptional repression using miRNA mimicking oligonucleotides or agomirs, is appropriate[97,98]. As a type of gene therapy, miRNA-based therapy has numerous advantages. First, the small molecular weight of miRNA oligonucleotides[98] renders them easier to deliver in vivo than plasmids or viral-based gene therapies. Second, miRNAs act as network regulators and can mediate silencing of multiple target genes or pathways simultaneously. This is believed to induce enhanced anti-cancer effects, compared to conventional therapies such as chemoradiotherapy and single-targeting inhibitors[97,99]. Third, miRNAs have lower toxicity compared with DNA or protein-based therapies because they are endogenously produced by cells[98,99]. Therefore, novel miRNA-based cancer therapies have become an exciting development in recent years.
Current studies on miRNAs in ESCC mostly focus on the identification of clinical correlation and novel targeting axes[100-102]. Less is known about the therapeutic potential of specific miRNAs in treatment. Isozaki et al[103] showed that subcutaneous injection of miR-375 suppressed ectopic ESCC tumor growth in vivo. Another miRNA, miR-27a, had similar tumor-suppressive effect on ESCC tumor growth in vivo after direct injection into implanted tumor[104]. Most recently, three miRNAs including miR-377, miR-29c, and miR-515-3p proved to have therapeutic potential through systemic delivery via intravenous injection in mouse models[38,40,74]. Specifically, systemically administered miR-377 was able to suppress the growth of subcutaneous ESCC tumor xenografts and lung metastasis in a mouse model[38]. Systemic delivery of miR-29c produced a synergistic effect in inhibiting tumor growth when combined with 5-FU treatment[40]. Oligonucleotide mimicking miR-515-3p inhibited lung metastasis of ESCC in nude mice[74].
CONCLUSION
Studies in recent years have underscored the salient involvement of miRNAs in cellular events and human disease. A number of dysregulated miRNAs have been identified in multiple cancer types and nominated as potential novel cancer biomarkers. We now have a better understanding of how miRNAs can simultaneously regulate multiple target genes, and thereby functionally behave as oncogenes or tumor suppressors during cancer development and progression. Most importantly, researchers have discovered the therapeutic potential of miRNAs as novel targets or agents. In summary, studies in ESCC confirmed that miRNAs form a complex regulatory network in modulating tumor progression and malignancy. However, related investigations on the clinical application of miRNAs are still scarce. Therefore, future in-depth research on regulatory mechanisms of miRNAs and further validation of their therapeutic efficacy will allow us to better exploit this ubiquitous class of small molecules.
Footnotes
Conflict-of-interest statement: The authors have no conflict of interest to declare.
Manuscript source: Invited manuscript
Peer-review started: January 27, 2021
First decision: May 7, 2021
Article in press: July 22, 2021
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim HS, Koritala T S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Guo X
Contributor Information
Di Cui, School of Biomedical Sciences, Li Ka Shing Faculty of Medicine, University of Hong Kong, Hong Kong 999077, China.
Annie LM Cheung, School of Biomedical Sciences, Li Ka Shing Faculty of Medicine, University of Hong Kong, Hong Kong 999077, China. lmcheung@hku.hk.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 3.Lin DC, Wang MR, Koeffler HP. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology. 2018;154:374–389. doi: 10.1053/j.gastro.2017.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 5.Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 6.Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112–120. doi: 10.4251/wjgo.v6.i5.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 8.Aguda BD. Modeling microRNA-transcription factor networks in cancer. Adv Exp Med Biol. 2013;774:149–167. doi: 10.1007/978-94-007-5590-1_9. [DOI] [PubMed] [Google Scholar]
- 9.Hemmatzadeh M, Mohammadi H, Karimi M, Musavishenas MH, Baradaran B. Differential role of microRNAs in the pathogenesis and treatment of Esophageal cancer. Biomed Pharmacother. 2016;82:509–519. doi: 10.1016/j.biopha.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Patnaik SK, Mallick R, Yendamuri S. MicroRNAs and esophageal cancer. J Gastrointest Oncol. 2010;1:55–63. doi: 10.3978/j.issn.2078-6891.2010.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan J, Wu W, Che Y, Kang N, Zhang R. Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Dis Esophagus. 2016;29:412–420. doi: 10.1111/dote.12338. [DOI] [PubMed] [Google Scholar]
- 12.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo X, Zhao L, Shen Y, Shao Y, Wei W, Liu F. Polymorphism of miRNA and esophageal cancer risk: an updated systemic review and meta-analysis. Onco Targets Ther. 2019;12:3565–3580. doi: 10.2147/OTT.S193958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen F, Chen J, Guo S, Zhou Y, Zheng Y, Yang Y, Zhang J, Wang X, Wang C, Zhao D, Wang M, Zhu M, Fan L, Xiang J, Xia Y, Wei Q, Jin L, Wang J. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol. 2016;37:4777–4784. doi: 10.1007/s13277-015-4268-3. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Vogelsang M, Schäfer G, Matejcic M, Parker MI. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PLoS One. 2013;8:e78520. doi: 10.1371/journal.pone.0078520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan Y, Weidhaas JB. Functional microRNA binding site variants. Mol Oncol. 2019;13:4–8. doi: 10.1002/1878-0261.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Li J, Guo H, Xu X, Xiong G, Guan X, Liu B, Chen X, Yang K, Bai Y. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33:2147–2154. doi: 10.1093/carcin/bgs259. [DOI] [PubMed] [Google Scholar]
- 18.Yao Y, Shao J, Wu J, Zhang Q, Wang J, Xiao D, Huang F. The Functional Variant in the 3'UTR of PTPRT with the Risk of Esophageal Squamous Cell Carcinoma in a Chinese Population. Cell Physiol Biochem. 2015;36:306–314. doi: 10.1159/000374073. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Su H, Hu N, Wang C, Wang L, Giffen C, Goldstein AM, Lee MP, Taylor PR. Integrated analysis of genome-wide miRNAs and targeted gene expression in esophageal squamous cell carcinoma (ESCC) and relation to prognosis. BMC Cancer. 2020;20:388. doi: 10.1186/s12885-020-06901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kano M, Seki N, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Peng J, Zhu W, Tao G, Song Y, Zhou X, Wang W. Combined downregulation of microRNA-133a and microRNA-133b predicts chemosensitivity of patients with esophageal squamous cell carcinoma undergoing paclitaxel-based chemotherapy. Med Oncol. 2014;31:263. doi: 10.1007/s12032-014-0263-6. [DOI] [PubMed] [Google Scholar]
- 22.Slotta-Huspenina J, Drecoll E, Feith M, Habermehl D, Combs S, Weichert W, Bettstetter M, Becker K, Langer R. MicroRNA expression profiling for the prediction of resistance to neoadjuvant radiochemotherapy in squamous cell carcinoma of the esophagus. J Transl Med. 2018;16:109. doi: 10.1186/s12967-018-1492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu N, Wang C, Clifford RJ, Yang HH, Su H, Wang L, Wang Y, Xu Y, Tang ZZ, Ding T, Zhang T, Goldstein AM, Giffen C, Lee MP, Taylor PR. Integrative genomics analysis of genes with biallelic loss and its relation to the expression of mRNA and micro-RNA in esophageal squamous cell carcinoma. BMC Genomics. 2015;16:732. doi: 10.1186/s12864-015-1919-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin HD, Liao XY, Chen YB, Huang SY, Xue WQ, Li FF, Ge XS, Liu DQ, Cai Q, Long J, Li XZ, Hu YZ, Zhang SD, Zhang LJ, Lehrman B, Scott AF, Lin D, Zeng YX, Shugart YY, Jia WH. Genomic Characterization of Esophageal Squamous Cell Carcinoma Reveals Critical Genes Underlying Tumorigenesis and Poor Prognosis. Am J Hum Genet. 2016;98:709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu YF, Yu JR, Yang Z, Zhu GX, Gao P, Wang H, Chen SY, Zhang J, Liu MY, Niu Y, Wei XM, Wang W, Ye FJ, Zhang LX, Zhao Y, Sun GG. Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC) J Exp Clin Cancer Res. 2018;37:301. doi: 10.1186/s13046-018-0966-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Gu J, Jiang P, Zheng Y, Liu X, Jiang X, Huang E, Xiong S, Xu F, Liu G, Ge D, Chu Y. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res. 2015;21:854–863. doi: 10.1158/1078-0432.CCR-14-1740. [DOI] [PubMed] [Google Scholar]
- 28.Yang M, Liu R, Li X, Liao J, Pu Y, Pan E, Wang Y, Yin L. Epigenetic Repression of miR-218 Promotes Esophageal Carcinogenesis by Targeting ROBO1. Int J Mol Sci. 2015;16:27781–27795. doi: 10.3390/ijms161126062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Guo T, Guo Y, Liu J, Qu F, He Y. Methylationassociated silencing of miR128 promotes the development of esophageal cancer by targeting COX2 in areas with a high incidence of esophageal cancer. Int J Oncol. 2019;54:644–654. doi: 10.3892/ijo.2018.4653. [DOI] [PubMed] [Google Scholar]
- 30.Pagano F, De Marinis E, Grignani F, Nervi C. Epigenetic role of miRNAs in normal and leukemic hematopoiesis. Epigenomics. 2013;5:539–552. doi: 10.2217/epi.13.55. [DOI] [PubMed] [Google Scholar]
- 31.Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, Andl CD. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015;14:24. doi: 10.1186/s12943-014-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang L, Zhao Z, Zheng L, Xue L, Zhan Q, Song Y. Downregulation of miR-503 Promotes ESCC Cell Proliferation, Migration, and Invasion by Targeting Cyclin D1. Genomics Proteomics Bioinformatics. 2017;15:208–217. doi: 10.1016/j.gpb.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng R, Liu Y, Zhang X, Zhao P, Deng Q. miRNA-200c enhances radiosensitivity of esophageal cancer by cell cycle arrest and targeting P21. Biomed Pharmacother. 2017;90:517–523. doi: 10.1016/j.biopha.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Ito T, Sato F, Kan T, Cheng Y, David S, Agarwal R, Paun BC, Jin Z, Olaru AV, Hamilton JP, Selaru FM, Yang J, Matsumura N, Shimizu K, Abraham JM, Shimada Y, Mori Y, Meltzer SJ. Polo-like kinase 1 regulates cell proliferation and is targeted by miR-593* in esophageal cancer. Int J Cancer. 2011;129:2134–2146. doi: 10.1002/ijc.25874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isozaki Y, Hoshino I, Nohata N, Kinoshita T, Akutsu Y, Hanari N, Mori M, Yoneyama Y, Akanuma N, Takeshita N, Maruyama T, Seki N, Nishino N, Yoshida M, Matsubara H. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int J Oncol. 2012;41:985–994. doi: 10.3892/ijo.2012.1537. [DOI] [PubMed] [Google Scholar]
- 36.Su X, Gao C, Feng X, Jiang M. miR-613 suppresses migration and invasion in esophageal squamous cell carcinoma via the targeting of G6PD. Exp Ther Med. 2020;19:3081–3089. doi: 10.3892/etm.2020.8540. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Ma J, Fan Y, Feng T, Chen F, Xu Z, Li S, Lin Q, He X, Shi W, Liu Y, Liu Z, Zhu B, Cao X. HOTAIR regulates HK2 expression by binding endogenous miR-125 and miR-143 in oesophageal squamous cell carcinoma progression. Oncotarget. 2017;8:86410–86422. doi: 10.18632/oncotarget.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, Law S, Xu LY, Li EM, Chan KW, Qin YR, Guan XY, He QY, Cheung ALM. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986–4000. doi: 10.1038/onc.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Z, Chen J, Chen X, Wang H, Tang L, Han C. microRNA-377 acts as a suppressor in esophageal squamous cell carcinoma through CBX3-dependent P53/P21 pathway. J Cell Physiol. 2021;236:107–120. doi: 10.1002/jcp.29631. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Hong P, Zheng CC, Dai W, Chen WY, Yang QS, Han L, Tsao SW, Chan KT, Lee NPY, Law S, Xu LY, Li EM, Chan KW, Qin YR, Guan XY, Lung ML, He QY, Xu WW, Cheung AL. Identification of miR-29c and its Target FBXO31 as a Key Regulatory Mechanism in Esophageal Cancer Chemoresistance: Functional Validation and Clinical Significance. Theranostics. 2019;9:1599–1613. doi: 10.7150/thno.30372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Han L, Li B, Yang J, Huen MS, Pan X, Tsao SW, Cheung AL. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of mitogen-activated protein kinase kinase 6 (MKK6) J Biol Chem. 2014;289:21508–21518. doi: 10.1074/jbc.M114.560342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang HL, Jiang Y, Wang YH, Chen T, He HJ, Liu T, Yang T, Yang LW, Chen J, Song ZQ, Yao W, Wu B, Liu G. FBXO31 promotes cell proliferation, metastasis and invasion in lung cancer. Am J Cancer Res. 2015;5:1814–1822. [PMC free article] [PubMed] [Google Scholar]
- 43.Han L, Cui D, Li B, Xu WW, Lam AKY, Chan KT, Zhu Y, Lee NPY, Law SYK, Guan XY, Qin YR, Chan KW, Ma S, Tsao SW, Cheung ALM. MicroRNA-338-5p reverses chemoresistance and inhibits invasion of esophageal squamous cell carcinoma cells by targeting Id-1. Cancer Sci. 2019;110:3677–3688. doi: 10.1111/cas.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin WC, Chen LH, Hsieh YC, Yang PW, Lai LC, Chuang EY, Lee JM, Tsai MH. miR-338-5p inhibits cell proliferation, colony formation, migration and cisplatin resistance in esophageal squamous cancer cells by targeting FERMT2. Carcinogenesis. 2019;40:883–892. doi: 10.1093/carcin/bgy189. [DOI] [PubMed] [Google Scholar]
- 45.Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. MiR-338-5p enhances the radiosensitivity of esophageal squamous cell carcinoma by inducing apoptosis through targeting survivin. Sci Rep. 2017;7:10932. doi: 10.1038/s41598-017-10977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen SW, Zhang YF, Li Y, Liu ZX, Lv HL, Li ZH, Xu YZ, Zhu YG, Tian ZQ. Characterization and effects of miR-21 expression in esophageal cancer. Genet Mol Res. 2015;14:8810–8818. doi: 10.4238/2015.August.3.4. [DOI] [PubMed] [Google Scholar]
- 47.Kimura S, Naganuma S, Susuki D, Hirono Y, Yamaguchi A, Fujieda S, Sano K, Itoh H. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep. 2010;23:1625–1633. doi: 10.3892/or_00000804. [DOI] [PubMed] [Google Scholar]
- 48.Nouraee N, Van Roosbroeck K, Vasei M, Semnani S, Samaei NM, Naghshvar F, Omidi AA, Calin GA, Mowla SJ. Expression, tissue distribution and function of miR-21 in esophageal squamous cell carcinoma. PLoS One. 2013;8:e73009. doi: 10.1371/journal.pone.0073009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komatsu S, Ichikawa D, Kawaguchi T, Miyamae M, Okajima W, Ohashi T, Imamura T, Kiuchi J, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating miR-21 as an independent predictive biomarker for chemoresistance in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6:1511–1523. [PMC free article] [PubMed] [Google Scholar]
- 50.Cai EH, Gao YX, Wei ZZ, Chen WY, Yu P, Li K. Serum miR-21 expression in human esophageal squamous cell carcinomas. Asian Pac J Cancer Prev. 2012;13:1563–1567. doi: 10.7314/apjcp.2012.13.4.1563. [DOI] [PubMed] [Google Scholar]
- 51.Komatsu S, Ichikawa D, Takeshita H, Konishi H, Nagata H, Hirajima S, Kawaguchi T, Arita T, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther. 2012;12 Suppl 1:S53–S59. doi: 10.1517/14712598.2012.681373. [DOI] [PubMed] [Google Scholar]
- 52.Liu F, Zheng S, Liu T, Liu Q, Liang M, Li X, Sheyhidin I, Lu X, Liu W. MicroRNA-21 promotes the proliferation and inhibits apoptosis in Eca109 via activating ERK1/2/MAPK pathway. Mol Cell Biochem. 2013;381:115–125. doi: 10.1007/s11010-013-1693-8. [DOI] [PubMed] [Google Scholar]
- 53.Wang N, Zhang CQ, He JH, Duan XF, Wang YY, Ji X, Zang WQ, Li M, Ma YY, Wang T, Zhao GQ. MiR-21 down-regulation suppresses cell growth, invasion and induces cell apoptosis by targeting FASL, TIMP3, and RECK genes in esophageal carcinoma. Dig Dis Sci. 2013;58:1863–1870. doi: 10.1007/s10620-013-2612-2. [DOI] [PubMed] [Google Scholar]
- 54.Liu T, Liu Q, Zheng S, Gao X, Lu M, Yang C, Dai F, Sheyhidin I, Lu X. MicroRNA-21 promotes cell growth and migration by targeting programmed cell death 4 gene in Kazakh's esophageal squamous cell carcinoma. Dis Markers. 2014;2014:232837. doi: 10.1155/2014/232837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H, Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, Sheyhidin I, Lu XM. Role of microRNA-21 and effect on PTEN in Kazakh's esophageal squamous cell carcinoma. Mol Biol Rep. 2011;38:3253–3260. doi: 10.1007/s11033-010-0480-9. [DOI] [PubMed] [Google Scholar]
- 56.Peng J, Lv Y, Wu C. Radiation-resistance increased by overexpression of microRNA-21 and inhibition of its target PTEN in esophageal squamous cell carcinoma. J Int Med Res. 2020;48:300060519882543. doi: 10.1177/0300060519882543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li P, Mao WM, Zheng ZG, Dong ZM, Ling ZQ. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Dig Dis Sci. 2013;58:3483–3493. doi: 10.1007/s10620-013-2854-z. [DOI] [PubMed] [Google Scholar]
- 58.Chen X, Cai S, Li B, Zhang X, Li W, Liang H, Cao X, Wang L, Wu Z. MicroRNA21 regulates the biological behavior of esophageal squamous cell carcinoma by targeting RASA1. Oncol Rep. 2019;41:1627–1637. doi: 10.3892/or.2018.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Z, Liu H, Shi Y, Yin L, Zhu Y, Liu R. Identification of Cancer Stem Cell Molecular Markers and Effects of hsa-miR-21-3p on Stemness in Esophageal Squamous Cell Carcinoma. Cancers (Basel) 2019;11 doi: 10.3390/cancers11040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 61.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 62.Fong LY, Taccioli C, Palamarchuk A, Tagliazucchi GM, Jing R, Smalley KJ, Fan S, Altemus J, Fiehn O, Huebner K, Farber JL, Croce CM. Abrogation of esophageal carcinoma development in miR-31 knockout rats. Proc Natl Acad Sci USA. 2020;117:6075–6085. doi: 10.1073/pnas.1920333117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu T, Ma P, Wu D, Shu Y, Gao W. Functions and mechanisms of microRNA-31 in human cancers. Biomed Pharmacother. 2018;108:1162–1169. doi: 10.1016/j.biopha.2018.09.132. [DOI] [PubMed] [Google Scholar]
- 64.Cui XB, Li S, Li TT, Peng H, Jin TT, Zhang SM, Liu CX, Yang L, Shen YY, Li SG, Li N, Li Y, Hu JM, Jiang JF, Suo J, Qi Y, Liang WH, Wang LH, Dang HW, Li L, Cao WW, Wei Y, Laibo-Yin , Wu CY, Yuan XL, Zhou H, Zheng Y, Chen YZ, Li F. Targeting oncogenic PLCE1 by miR-145 impairs tumor proliferation and metastasis of esophageal squamous cell carcinoma. Oncotarget. 2016;7:1777–1795. doi: 10.18632/oncotarget.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao L, Wei ZB, Yang CQ, Chen JJ, Li D, Ji AF, Ma L. Effects of PLCE1 gene silencing by RNA interference on cell cycling and apoptosis in esophageal carcinoma cells. Asian Pac J Cancer Prev. 2014;15:5437–5442. doi: 10.7314/apjcp.2014.15.13.5437. [DOI] [PubMed] [Google Scholar]
- 66.Yu S, Choi WI, Choi YJ, Kim HY, Hildebrandt F, Gee HY. PLCE1 regulates the migration, proliferation, and differentiation of podocytes. Exp Mol Med. 2020;52:594–603. doi: 10.1038/s12276-020-0410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang HF, Zhang K, Liao LD, Li LY, Du ZP, Wu BL, Wu JY, Xu XE, Zeng FM, Chen B, Cao HH, Zhu MX, Dai LH, Long L, Wu ZY, Lai R, Xu LY, Li EM. miR-200b suppresses invasiveness and modulates the cytoskeletal and adhesive machinery in esophageal squamous cell carcinoma cells via targeting Kindlin-2. Carcinogenesis. 2014;35:292–301. doi: 10.1093/carcin/bgt320. [DOI] [PubMed] [Google Scholar]
- 68.Paolillo M, Schinelli S. Extracellular Matrix Alterations in Metastatic Processes. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20194947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, Ma J, Guo Q, Duan F, Tang F, Zheng P, Zhao Z, Lu G. Overexpression of MMP-2 and MMP-9 in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:664–667. doi: 10.1111/j.1442-2050.2008.00928.x. [DOI] [PubMed] [Google Scholar]
- 70.Guan X, Wang X, Luo H, Wu J, Zhang X. Matrix metalloproteinase 1, 3, and 9 polymorphisms and esophageal squamous cell carcinoma risk. Med Sci Monit. 2014;20:2269–2274. doi: 10.12659/MSM.892413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang L, Song X, Zhu J, Li M, Ji Y, Wu F, Chen Y, Cui X, Hu J, Wang L, Cao Y, Wei Y, Zhang W, Li F. Tumor suppressor microRNA-34a inhibits cell migration and invasion by targeting MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J Oncol. 2017;51:378–388. doi: 10.3892/ijo.2017.4015. [DOI] [PubMed] [Google Scholar]
- 72.Nie J, Ge X, Geng Y, Cao H, Zhu W, Jiao Y, Wu J, Zhou J, Cao J. miR-34a inhibits the migration and invasion of esophageal squamous cell carcinoma by targeting Yin Yang-1. Oncol Rep. 2015;34:311–317. doi: 10.3892/or.2015.3962. [DOI] [PubMed] [Google Scholar]
- 73.Qi Y, Li X, Zhao S. miR-29b inhibits the progression of esophageal squamous cell carcinoma by targeting MMP-2. Neoplasma. 2015;62:384–390. doi: 10.4149/neo_2015_046. [DOI] [PubMed] [Google Scholar]
- 74.Hu HF, Xu WW, Zhang WX, Yan X, Li YJ, Li B, He QY. Identification of miR-515-3p and its targets, vimentin and MMP3, as a key regulatory mechanism in esophageal cancer metastasis: functional and clinical significance. Signal Transduct Target Ther. 2020;5:271. doi: 10.1038/s41392-020-00275-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osako Y, Seki N, Kita Y, Yonemori K, Koshizuka K, Kurozumi A, Omoto I, Sasaki K, Uchikado Y, Kurahara H, Maemura K, Natsugoe S. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int J Oncol. 2016;49:2255–2264. doi: 10.3892/ijo.2016.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akanuma N, Hoshino I, Akutsu Y, Murakami K, Isozaki Y, Maruyama T, Yusup G, Qin W, Toyozumi T, Takahashi M, Suito H, Hu X, Sekino N, Matsubara H. MicroRNA-133a regulates the mRNAs of two invadopodia-related proteins, FSCN1 and MMP14, in esophageal cancer. Br J Cancer. 2014;110:189–198. doi: 10.1038/bjc.2013.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsubakihara Y, Moustakas A. Epithelial-Mesenchymal Transition and Metastasis under the Control of Transforming Growth Factor β. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian X, Fei Q, Du M, Zhu H, Ye J, Qian L, Lu Z, Zhang W, Wang Y, Peng F, Chen J, Liu B, Li Q, He X, Yin L. miR-130a-3p regulated TGF-β1-induced epithelial-mesenchymal transition depends on SMAD4 in EC-1 cells. Cancer Med. 2019;8:1197–1208. doi: 10.1002/cam4.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang F, Wang J, Yang X, Chen D, Wang L. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 2016;11:88. doi: 10.1186/s13000-016-0536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dai F, Liu T, Zheng S, Liu Q, Yang C, Zhou J, Chen Y, Sheyhidin I, Lu X. MiR-106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh's esophageal squamous cell carcinoma. Tumour Biol. 2016;37:14595–14604. doi: 10.1007/s13277-016-5338-x. [DOI] [PubMed] [Google Scholar]
- 85.Chang HY, Lee CH, Li YS, Huang JT, Lan SH, Wang YF, Lai WW, Wang YC, Lin YJ, Liu HS, Cheng HC. MicroRNA-146a suppresses tumor malignancy via targeting vimentin in esophageal squamous cell carcinoma cells with lower fibronectin membrane assembly. J Biomed Sci. 2020;27:102. doi: 10.1186/s12929-020-00693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma T, Zhao Y, Lu Q, Lu Y, Liu Z, Xue T, Shao Y. MicroRNA-30c functions as a tumor suppressor via targeting SNAI1 in esophageal squamous cell carcinoma. Biomed Pharmacother. 2018;98:680–686. doi: 10.1016/j.biopha.2017.12.095. [DOI] [PubMed] [Google Scholar]
- 87.Zong M, Liu Y, Zhang K, J Y, Chen L. The effects of miR-429 on cell migration and invasion by targeting Slug in esophageal squamous cell carcinoma. Pathol Res Pract. 2019;215:152526. doi: 10.1016/j.prp.2019.152526. [DOI] [PubMed] [Google Scholar]
- 88.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao L, Li R, Xu S, Li Y, Zhao P, Dong W, Liu Z, Zhao Q, Tan B. Tumor suppressor miR-128-3p inhibits metastasis and epithelial-mesenchymal transition by targeting ZEB1 in esophageal squamous-cell cancer. Acta Biochim Biophys Sin (Shanghai) 2018;50:171–180. doi: 10.1093/abbs/gmx132. [DOI] [PubMed] [Google Scholar]
- 90.Yokobori T, Suzuki S, Tanaka N, Inose T, Sohda M, Sano A, Sakai M, Nakajima M, Miyazaki T, Kato H, Kuwano H. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013;104:48–54. doi: 10.1111/cas.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z, Sun J, Tan FW, Ding DP, Xu XH, Zhou F, Tan XG, Hang J, Shi SS, Feng XL, He J. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J Biol Chem. 2011;286:10725–10734. doi: 10.1074/jbc.M110.165654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu B, Li X, Li C, Xu R, Sun X. miR-25 mediates metastasis and epithelial-mesenchymal-transition in human esophageal squamous cell carcinoma via regulation of E-cadherin signaling. Bioengineered. 2019;10:679–688. doi: 10.1080/21655979.2019.1687391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen F, Chu L, Li J, Shi Y, Xu B, Gu J, Yao X, Tian M, Yang X, Sun X. Hypoxia induced changes in miRNAs and their target mRNAs in extracellular vesicles of esophageal squamous cancer cells. Thorac Cancer. 2020;11:570–580. doi: 10.1111/1759-7714.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu WW, Li B, Guan XY, Chung SK, Wang Y, Yip YL, Law SY, Chan KT, Lee NP, Chan KW, Xu LY, Li EM, Tsao SW, He QY, Cheung AL. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat Commun. 2017;8:14399. doi: 10.1038/ncomms14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang W, Hong R, Li L, Wang Y, Du P, Ou Y, Zhao Z, Liu X, Xiao W, Dong D, Wu Q, Chen J, Song Y, Zhan Q. The chromosome 11q13.3 amplification associated lymph node metastasis is driven by miR-548k through modulating tumor microenvironment. Mol Cancer. 2018;17:125. doi: 10.1186/s12943-018-0871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu Y, Chen J, Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA. 2019;1:24. [Google Scholar]
- 98.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Myoung S, Kasinski AL. Chapter 14: Strategies for safe and targeted delivery of microRNA therapeutics. In: MicroRNAs in Diseases and Disorders. Emerging Therapeutic Targets: The Royal Society of Chemistry, 2019: 386-415. [Google Scholar]
- 100.Sakai NS, Samia-Aly E, Barbera M, Fitzgerald RC. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin Cancer Biol. 2013;23:512–521. doi: 10.1016/j.semcancer.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 101.Harada K, Baba Y, Ishimoto T, Shigaki H, Kosumi K, Yoshida N, Watanabe M, Baba H. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–530. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 102.Mei LL, Qiu YT, Zhang B, Shi ZZ. MicroRNAs in esophageal squamous cell carcinoma: Potential biomarkers and therapeutic targets. Cancer Biomark. 2017;19:1–9. doi: 10.3233/CBM-160240. [DOI] [PubMed] [Google Scholar]
- 103.Isozaki Y, Hoshino I, Akutsu Y, Hanari N, Mori M, Nishimori T, Murakami K, Akanuma N, Takeshita N, Maruyama T, Toyozumi T, Takahashi M, Suito H, Matsubara H. Usefulness of microRNA375 as a prognostic and therapeutic tool in esophageal squamous cell carcinoma. Int J Oncol. 2015;46:1059–1066. doi: 10.3892/ijo.2014.2789. [DOI] [PubMed] [Google Scholar]
- 104.Wang X, An D, Liu X, Wang X, Li B. MicroRNA-27a downregulates the expression of Hsp90 and enhances the radiosensitivity in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:5967–5977. doi: 10.2147/OTT.S197456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu W, Li M, Chen X, Zhu S, Shi H, Zhang D, Cheng C, Li B. MicroRNA-1 suppresses proliferation, migration and invasion by targeting Notch2 in esophageal squamous cell carcinoma. Sci Rep. 2018;8:5183. doi: 10.1038/s41598-018-23421-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Q, Zhang J, Fu Z, Dong L, Tang Y, Xu C, Wang H, Zhang T, Wu Y, Dong C, Shao S, Wang G. Hypoxia-induced microRNA-10b-3p promotes esophageal squamous cell carcinoma growth and metastasis by targeting TSGA10. Aging (Albany NY) 2019;11:10374–10384. doi: 10.18632/aging.102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jing C, Ma G, Li X, Wu X, Huang F, Liu K, Liu Z. MicroRNA-17/20a impedes migration and invasion via TGF-β/ITGB6 pathway in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6:1549–1562. [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y, Pan T, Zhong X, Cheng C. Nicotine upregulates microRNA-21 and promotes TGF-β-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014;35:7063–7072. doi: 10.1007/s13277-014-1968-z. [DOI] [PubMed] [Google Scholar]
- 109.Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D, Wang L, Wang YJ, Zong Y, Wu YD, Zhang ST. MiR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget. 2016;7:15173–15186. doi: 10.18632/oncotarget.7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zuo J, Zhu K, Wang Y, Yu Z. MicroRNA-34a suppresses invasion and metastatic in esophageal squamous cell carcinoma by regulating CD44. Mol Cell Biochem. 2018;443:139–149. doi: 10.1007/s11010-017-3218-3. [DOI] [PubMed] [Google Scholar]
- 111.Ma G, Jing C, Li L, Huang F, Ding F, Wang B, Lin D, Luo A, Liu Z. MicroRNA-92b represses invasion-metastasis cascade of esophageal squamous cell carcinoma. Oncotarget. 2016;7:20209–20222. doi: 10.18632/oncotarget.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang S, Zhang L, Shi P, Zhang Y, Zhou H, Cao X. Genome-wide profiles of metastasis-associated mRNAs and microRNAs in salivary adenoid cystic carcinoma. Biochem Biophys Res Commun. 2018;500:632–638. doi: 10.1016/j.bbrc.2018.04.122. [DOI] [PubMed] [Google Scholar]
- 113.Wang W, Wu D, He X, Hu X, Hu C, Shen Z, Lin J, Pan Z, He Z, Lin H, Wang M. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi: 10.1016/j.canlet.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 114.Wang T, Chen T, Niu H, Li C, Xu C, Li Y, Huang R, Zhao J, Wu S. MicroRNA-218 inhibits the proliferation and metastasis of esophageal squamous cell carcinoma cells by targeting BMI1. Int J Mol Med. 2015;36:93–102. doi: 10.3892/ijmm.2015.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang B, Zhang Z, Li L, Qin YR, Liu H, Jiang C, Zeng TT, Li MQ, Xie D, Li Y, Guan XY, Zhu YH. TSPAN15 interacts with BTRC to promote oesophageal squamous cell carcinoma metastasis via activating NF-κB signaling. Nat Commun. 2018;9:1423. doi: 10.1038/s41467-018-03716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yi J, Jin L, Chen J, Feng B, He Z, Chen L, Song H. MiR-375 suppresses invasion and metastasis by direct targeting of SHOX2 in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:159–169. doi: 10.1093/abbs/gmw131. [DOI] [PubMed] [Google Scholar]
- 117.Jin L, Yi J, Gao Y, Han S, He Z, Chen L, Song H. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) 2016;48:810–819. doi: 10.1093/abbs/gmw073. [DOI] [PubMed] [Google Scholar]
- 118.Zhang JX, Chen ZH, Xu Y, Chen JW, Weng HW, Yun M, Zheng ZS, Chen C, Wu BL, Li EM, Fu JH, Ye S, Xie D. Downregulation of MicroRNA-644a Promotes Esophageal Squamous Cell Carcinoma Aggressiveness and Stem Cell-like Phenotype via Dysregulation of PITX2. Clin Cancer Res. 2017;23:298–310. doi: 10.1158/1078-0432.CCR-16-0414. [DOI] [PubMed] [Google Scholar]
- 119.Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol. 2015;21:3245–3255. doi: 10.3748/wjg.v21.i11.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou J, Zhu J, Jiang G, Feng J, Wang Q. Downregulation of microRNA-4324 promotes the EMT of esophageal squamous-cell carcinoma cells via upregulating FAK. Onco Targets Ther. 2019;12:4595–4604. doi: 10.2147/OTT.S198333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meng L, Liu F, Ju Y, Ding P, Liu S, Chang S, Zhang Y, Lian Y, Gu L, Zhang X, Sang M. Tumor suppressive miR-6775-3p inhibits ESCC progression through forming a positive feedback loop with p53 via MAGE-A family proteins. Cell Death Dis. 2018;9:1057. doi: 10.1038/s41419-018-1119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]