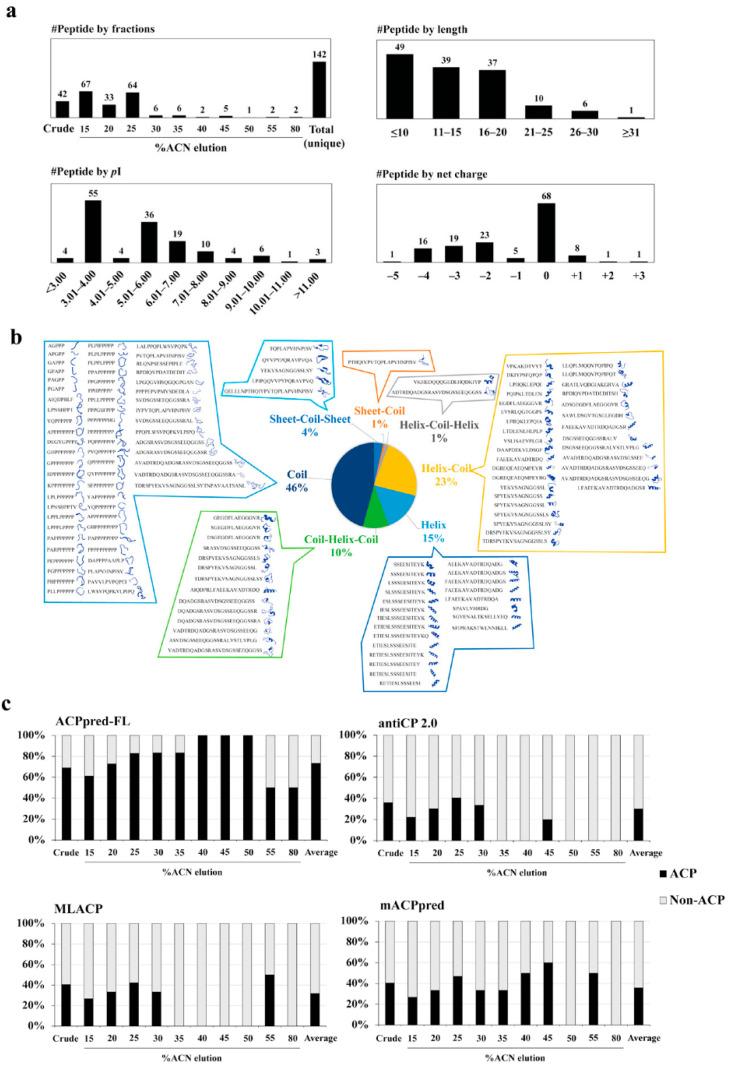
Figure 3.
Predicted physicochemical, structural, and machine learning-based anticancer peptide screening of naturally occurring human milk peptides. (a) Distribution of the unique human milk peptides identified by LC-MS/MS; (b) the distribution and secondary structure of all identified peptides predicted by PEP-FOLD3 software; (c) proportions of predicted anticancer property (ACP) vs. non-ACP peptides using four ACP machine learning programs, including ACPpred-FL, antiCP 2.0, MLACP, and mACPpred. The percentages of ACP (black) and non-ACP (gray) peptides were calculated as: number of ACP or non-ACP predictions/number of total identified peptides in each individual fraction × 100%. Full results of in silico ACP screening are provided in Table S2.