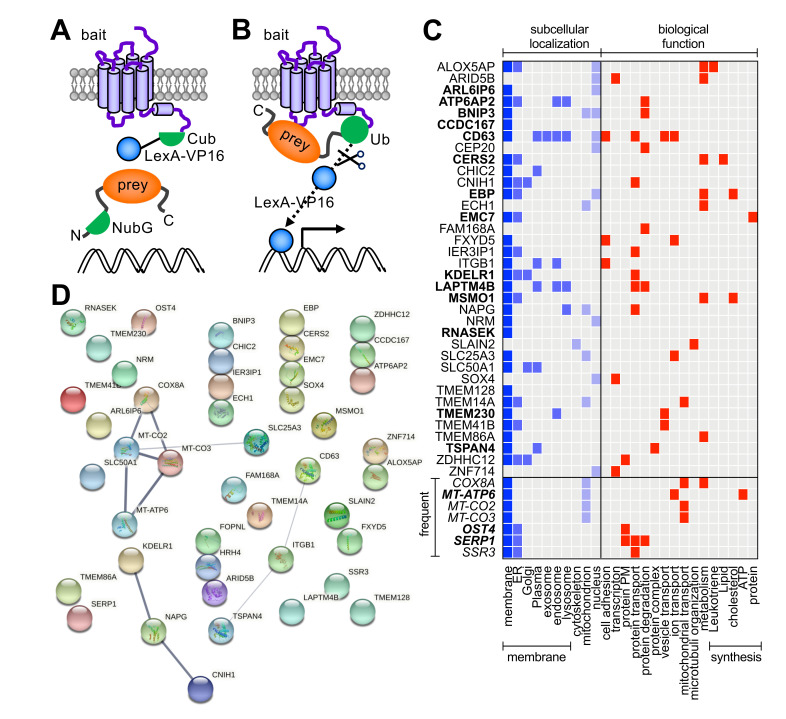
Figure 1.
Schematic overview of the split-ubiquitin membrane-based yeast two-hybrid (MYTH) assay. (A) Diploid yeast co-expressing the membrane targeted H4R-Cub-LexA-VP16 bait and cytosolic (depicted) or membrane-associated N-terminally tagged NubG prey proteins was generated by mating of haploid yeasts expressing the individual bait and prey constructs. (B) Interaction of the H4R-Cub-LexA-VP16 ‘bait’ with NubG-protein ‘prey’ results in functional reconstitution of a pseudo-ubiquitin, which is subsequently recognized by cytosolic ubiquitin-specific proteases leading to the cleavage of the LexA-VP16 transcriptional activator and the expression of (HIS3, ADE2, LacZ) reporter genes. (C) Subcellular localization and biological function summary of the 43 hits were retrieved from the Uniprot database (https://www.uniprot.org; accessed on 24 June 2021). Gene names are indicated and full description of proteins with Uniprot codes are presented in Supplementary Table S1. Hits in bold have been found to interact with other GPCRs in MYTH screens (see Supplementary Table S1), whereas frequent MYTH screen hits are indicated in italics. ER = endoplasmic reticulum; PM = post-translational modification (D) STRING analysis (v.11.0; https://string-db.org; accessed on 25 June 2021) of hits from the MYTH screen of H4R (bait) on an unstimulated Jurkat T cell DUALmembrane cDNA library. Known interactions between proteins in the STRING database are indicated by connecting lines. The thickness of the line represents the degree of confidence prediction of the interaction.