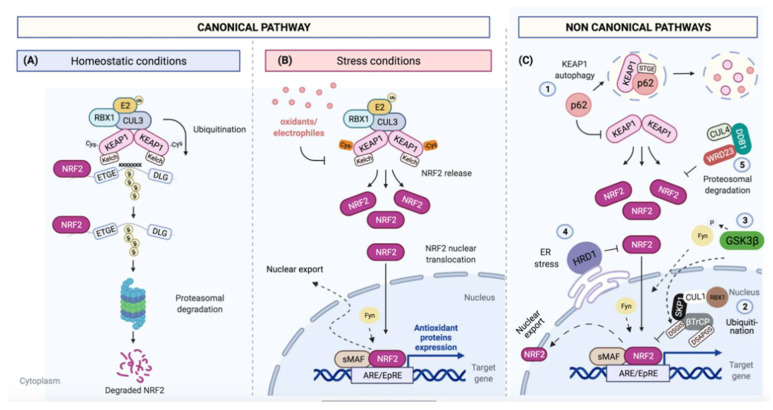
Figure 3.
Canonical and non-canonical NRF2 pathways. (A) Under basal conditions, NRF2 ETGE/DLG motifs bind to KEAP1 Kelch domains. Binding to KEAP1 brings the CUL3/RBX1 E3 ubiquitin ligase into the complex and targets NRF2 for poly-ubiquitination and degradation by 26S proteasome; (B) Several oxidative and electrophilic stressors can modify critical KEAP1 cysteine residues, disrupting the KEAP1-NRF2 complex. As a consequence, NRF2 protein levels increase, causing its translocation into the nucleus where it forms a heterodimer with sMAF transcriptional factors to act on ARE/EpRE enhancer sequences for the control of its transcriptional program. Afterwards, NRF2 is exported to cytoplasm upon phosphorylation by different Src family kinases, such as Fyn; (C) NRF2 can be also regulated by KEAP1-independent mechanisms: (1) by p62/SQSTM1, which promotes KEAP1 autophagic degradation via its STGE binding motif; (2) by β-TrCP that can form a complex with CUL1/SKP1, promoting NRF2 ubiquitination and degradation; (3) by GSK-3β, which can phosphorylate β-TrCP, increasing NRF2 ubiquitination; (4) by HRD1, which can interact under reticulum stress conditions with Neh4 and 5 domains and trigger NRF2. Recently, (5) CUL4/DDB1/WDR23 was discovered as another E3 ubiquitin ligase to regulate NRF2, but its mechanism is still unclear.