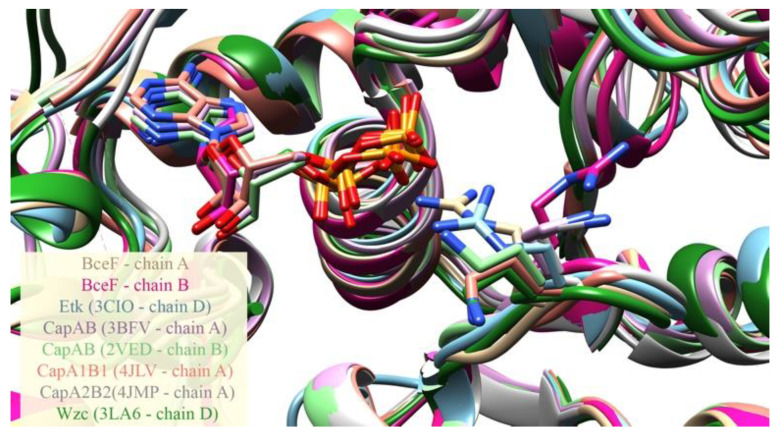
Figure 2.
Conformational variability of the ADP-proximal residue Arg590 in BceF and its equivalent positions in other BY-kinases. ADP is present in both BceF chains, and CapAB (PDB Ids 3BFV, 2VED and 4JLV). ADP carbons are colored the same as the backbone of the protein from the same determined structure. The figure depicts a structural superimposition of the kinase domain of several BY-kinases, and of the two chains in the asymmetric unit in the crystal structure of BceF, with zoom-in into the ADP ligand and into the Arg590 residue in BceF and the equivalent positions in the other BY-kinases, shown as sticks. Side-chain atoms are colored by atom types (nitrogen in blue, oxygen in red and sulfur in yellow). The backbone and carbon atoms in each BY-kinase and BceF chain are colored differently: BceF chain A in beige, BceF chain B in magenta, Etk (PDB code 3CIO) in cyan (featuring the equivalent Arg572), CapAB (PDB code 3BFV in purple, PDB code 2VED in light green, PDB code 4JLV in orange, PDB code 4JMP in grey), featuring the equivalent Lys1082, and Wzc (PDB code 3LA6 in dark green), featuring the equivalent Lys567.