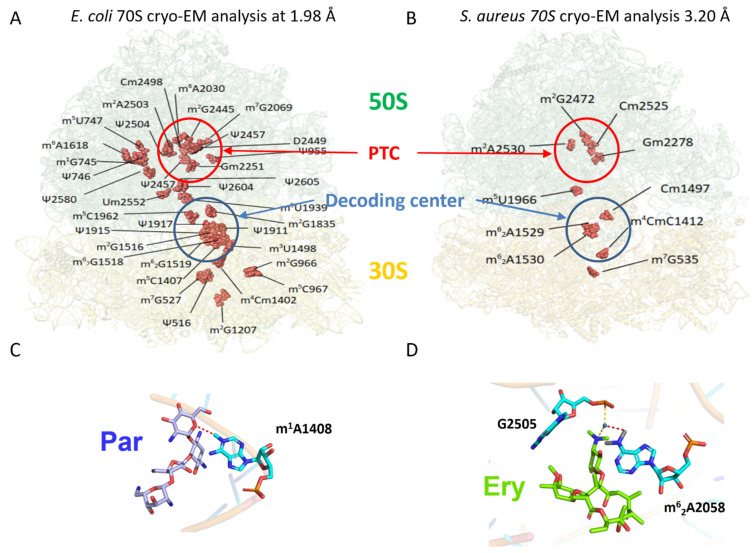
Figure 1.
Natural ribosomal RNA modifications and additional modifications implicated in antibiotic resistance mechanisms as visualized by structural analyses. (A) Cryo-EM structure at 1.98 Å of E. coli 70S ribosome (pdb file 7K00). 11 and 24 RNA modifications (red spheres) could be visualized in the 30S (16S rRNA) and 50S (23S rRNA) subunits, respectively [88]. (B) Cryo-EM structure at 3.20 Å of S. aureus 70S ribosome (pdb file 6YEF). The limited resolution allowed the visualization of 4 modifications in the 16S rRNA and 6 in 23S rRNA [89]. PTC, Peptidyl Transferase Center on the large 50S subunit. (C) Mechanism of aminoglycoside (Par, paromomycin) resistance induced by methylation of A1408 (pdb file 5ZEJ [90]). The presence of the methyl group directly perturbs antibiotic interaction. (D) Mechanism of macrolide (Ery, erythromycin) resistance induced by dimethylation of A2058 (pdb file 6XHV [91]). The two methyl groups on A2058 prevent the coordination of a water molecule with G2505, which stabilizes erythromycin binding.