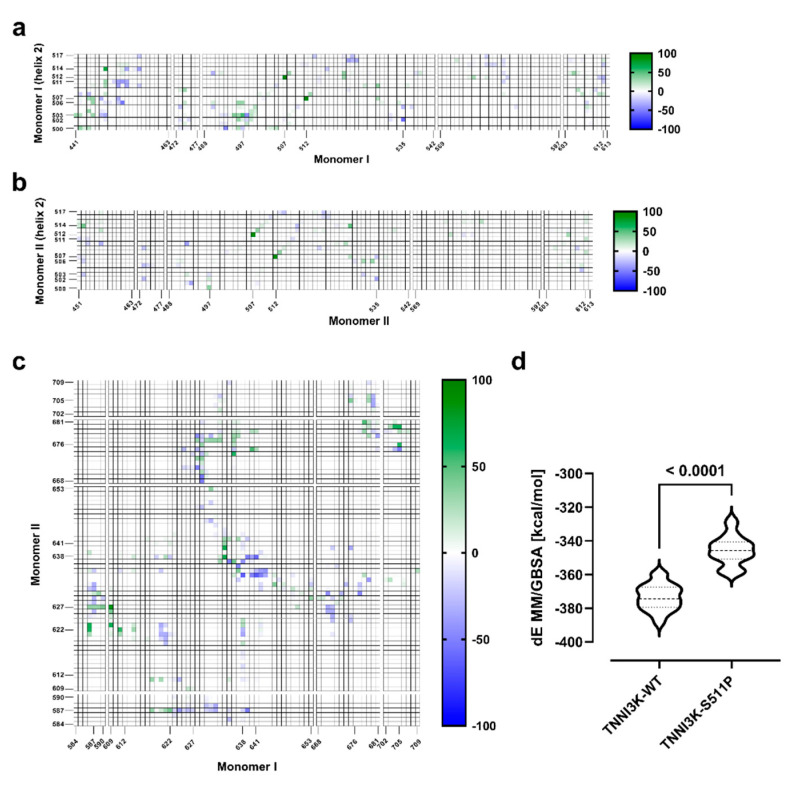
Figure 3.
Average intra/inter residue-residue contact maps and MM/GBSA analysis. (a) Contact map of the residues of helix 2 in monomer I and the residues of the corresponding monomer, and (b) residues of helix 2 in monomer II and the residues of the corresponding monomer. Each point represents the mean of the average residue-residue contact difference between TNNI3K-WT and TNNI3K-S511P over the last 200 ns from the three independent MD simulations (spectrum range from green to white to blue, from 100% to −100% contact; green means more contacts in TNNI3K-WT and blue means more contacts in TNNI3K-S511P). (c) Contact map between residue-residue pairs in the dimer interface for TNNI3K-WT vs. TNNI3K-S511P. The contact spectrum ranging from green to white to blue, representing 100% to −100% contact, where green indicates more contacts in TNNI3K-WT and blue indicates more contacts in TNNI3K-S511P. Each point represents the mean difference in the residue-residue contact over the last 200 ns from the three independent MD simulations. (d) MM/GBSA analysis applied for dimerization of TNNI3K. Every 10th snapshot of the last 200 ns of the MD simulation was used. Each violin plot represents the mean from the three independent simulations (95% confidence interval).