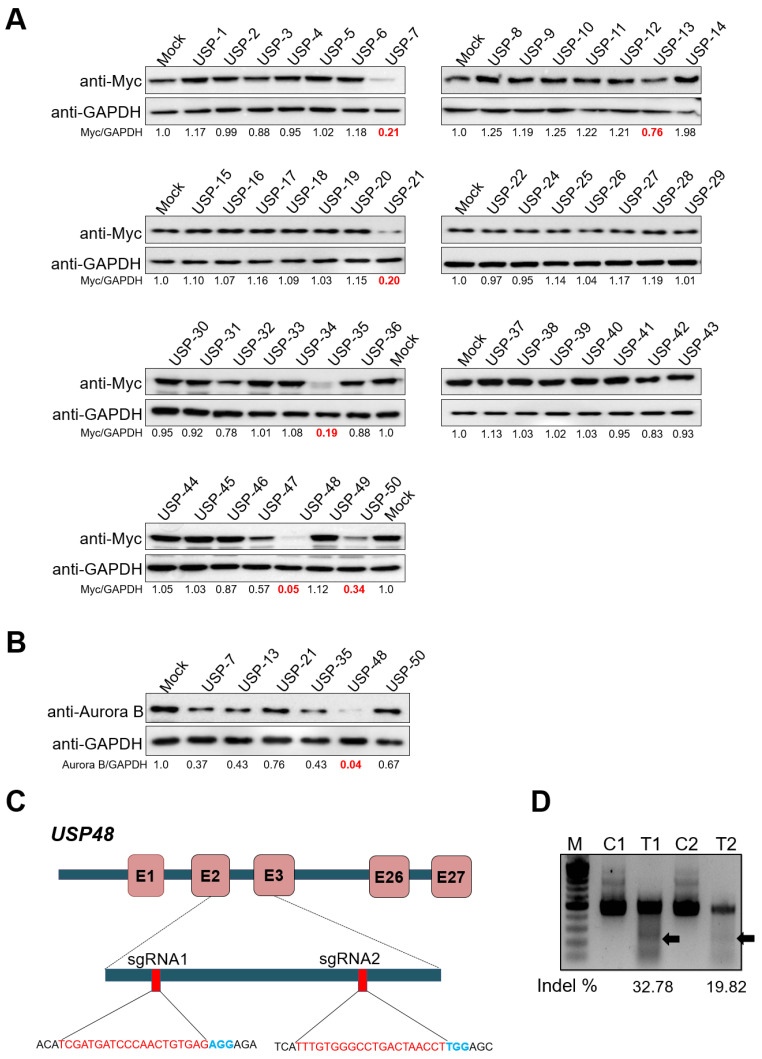
Figure 1.
CRISPR-based genome-scale screening of USP sub-family proteins exhibiting a reduction in Aurora B protein levels. (A) Screening for DUBs regulating Myc-Aurora B was performed using a CRISPR-Cas9-based DUBKO sgRNA kit. HEK293 cells were transfected with Myc-Aurora B and the indicated sgRNAs along with Cas9. Equal concentrations of proteins from DUBKO HEK293 cell lysates were subjected to Western blot analyses. The protein band intensities were estimated using ImageJ software with reference to the GAPDH control band for each individual sgRNA (Myc-Aurora B/GAPDH). The loss of USPs leading to the downregulation of the Aurora B protein level is marked in red. (B) The putative DUBs USP7, USP13, USP21, USP35, USP48, and USP50, which potentially regulate Aurora B protein levels, were transfected in HeLa cells along with Cas9. The protein band intensities were estimated using ImageJ software with reference to the GAPDH control band for each individual sgRNA (Aurora B/GAPDH). The protein band intensity of HeLa cells transfected with sgRNA targeting USP48 showed the highest reduction in the endogenous Aurora B is represented in red. (C) Schematic of RNA-guided engineered nuclease targeting the sequences in exon 2 and exon 3 of the human USP48 gene using sgRNA1 and sgRNA2, respectively. PAM sequences are represented in blue, while the sgRNA target sequences are represented in red. (D) T7E1 assays were performed in HEK293 cells to determine the cleavage efficiency of sgRNA1 (T1) and sgRNA2 (T2). The cleaved band intensity (indicated by arrow) obtained from the T7E1 assay was estimated using ImageJ software and represented as indel percentage (indel %). Scrambled sgRNA-transfected cells were used as control cells (C). A marker is shown for size reference.