Figure 2.
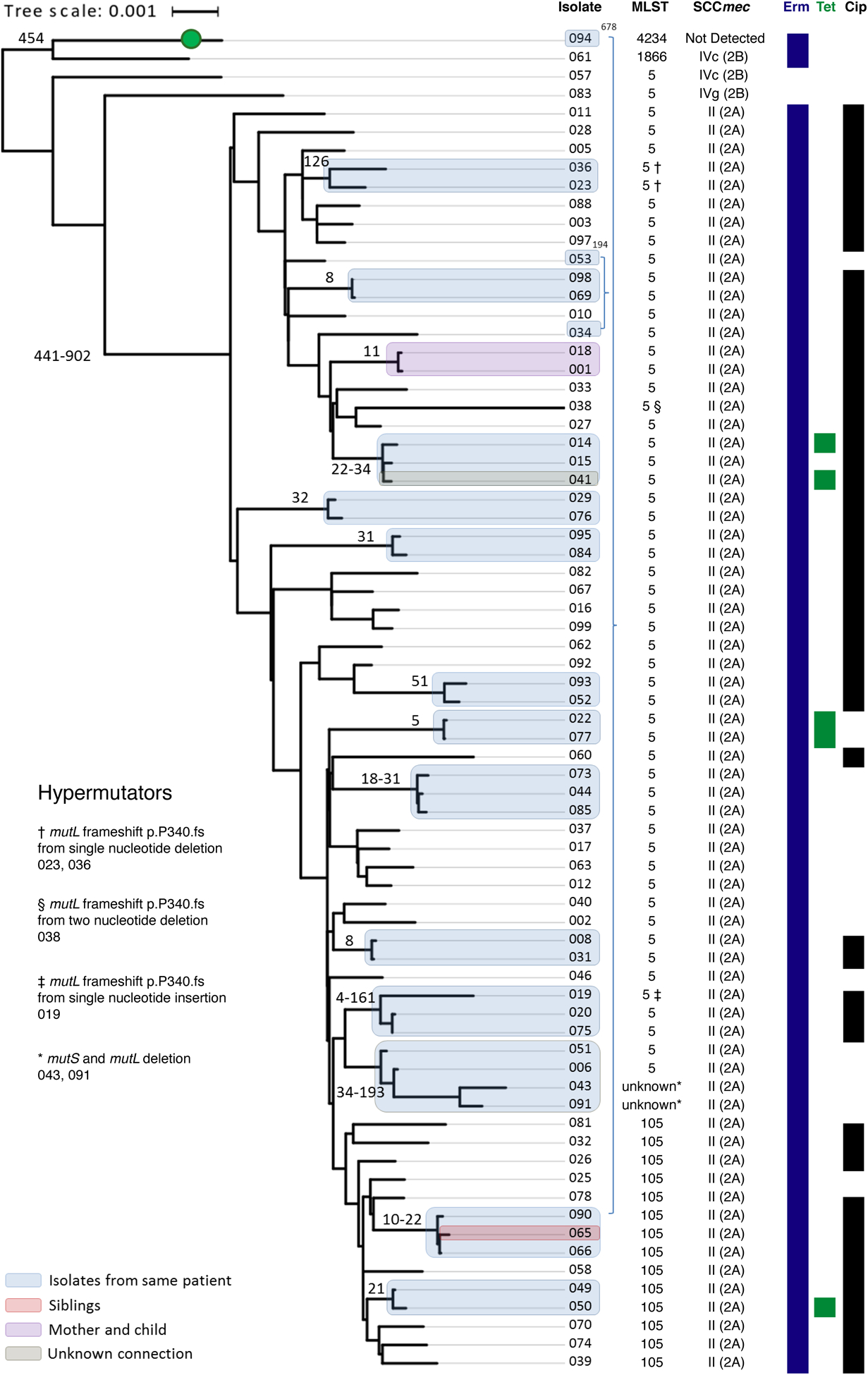
Lyve-SET Phylogenetic reconstruction for clade 1 reveals closely related strains within subjects or shared by different individuals. Sequence type (ST), SCCmec element, and predicted antibiotic resistance from genome sequence are given at right. A green circle indicates an isolate predicted to be MSSA based on lack of detection of SCCmec. Antibiotic susceptibilities denote isolates predicted to be resistant to erythromycin (Erm), tetracycline (Tet), or ciprofloxacin (Cip). Highly related isolates are indicated with shaded boxes, colored based on the relationship between patients. The SNP difference between these isolates is given as a number adjacent to the shaded box. Several isolates with increased genetic distance were found to have mutations in mutL and/or mutS. Tree scale represents substitutions per site
