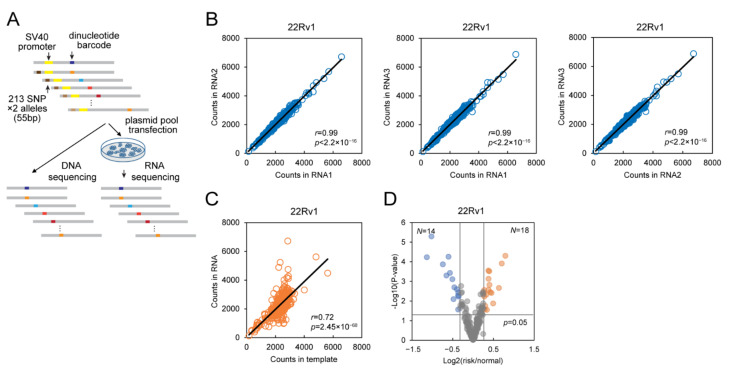
Figure 1.
DiR-seq assays discovered regulatory SNPs in prostate cancer cells. (A) Flowchart of the DiR system for reporter gene assay in prostate cancer cells. The DiR plasmid library was developed based on the pGL3-promoter vector by optimizing the luciferase coding sequence. The 55 bp SNPs fragment was inserted upstream of the SV40 promoter, 450 bp dinucleotide-barcoded sequences were used as the reporter gene. (B) Consistency evaluation between individual biological replicates of DiR-seq assay of 213 prostate cancer risk SNPs in 22Rv1 cells. The correlation coefficient values and p values were calculated with Pearson correlation analysis (C) Scatter plot of DiR-seq tag counts in RNA and DNA template in 22Rv1 cells. The correlation coefficient values and p values were calculated with Pearson correlation analysis. (D) Volcano plot of the allelic ratio of reporter expression levels in DiR-seq analysis in the 22Rv1 cell line. p values came from a two-tailed Student’s t-test of the reporter expression of individual alleles. The blue dots represent SNP sites satisfying the criteria of fold change < 0.8, p < 0.05, and the orange dots represent SNP sites satisfying the criteria of fold change > 1.2, p < 0.05.