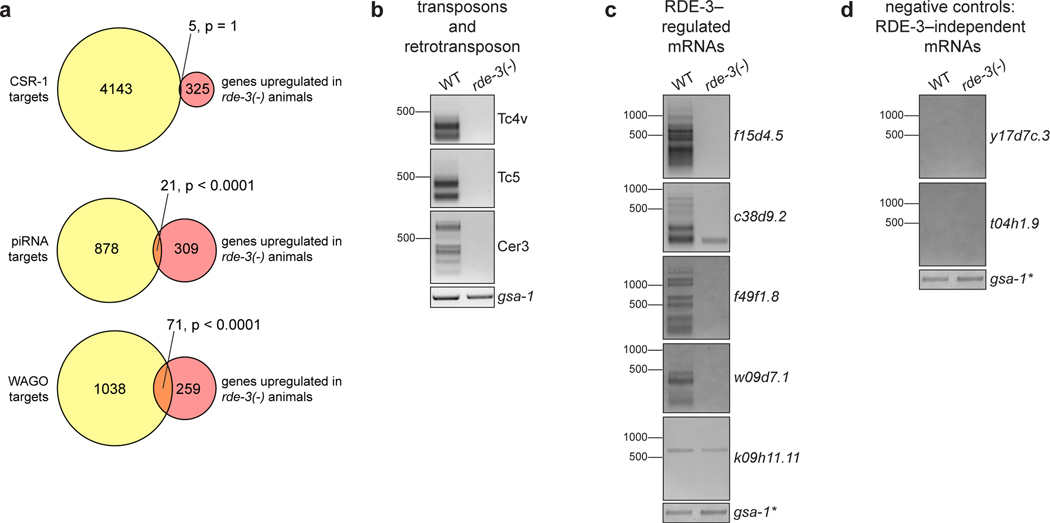
Extended Data Fig. 5. Endogenous targets of pUGylation in C. elegans.
a, mRNAs upregulated in rde-3(−) mutants (Supplementary Table 2) were compared to published lists of: (1) RNAs targeted by CSR-1–bound endo-siRNAs52, (2) piRNA-targeted mRNAs (based on predictive and experimental approaches)53, and (3) WAGO-class mRNAs26. p-values were generated using a one-sided Fisher’s exact test. This analysis showed statistically significant overlap between the mRNAs upregulated in rde-3(−) mutants and both piRNAs targets and WAGO-class mRNAs. b-d, Total RNA was extracted from WT or rde-3(−) animals. The assay outlined in Fig. 1a was used to detect pUG RNAs for b, two DNA transposons (Tc4v and Tc5) and a retrotransposon (Cer3) that were significantly upregulated in rde-3(−) animals; c, predicted protein-coding mRNAs that were significantly upregulated in rde-3(−) animals; and d, two randomly selected mRNAs whose expression does not change in rde-3(−) mutants. Data is representative of 3 biologically independent experiments. *Note: the same RT samples were used for panels c and d and, therefore, the gsa-1 loading control is the same for both panels.