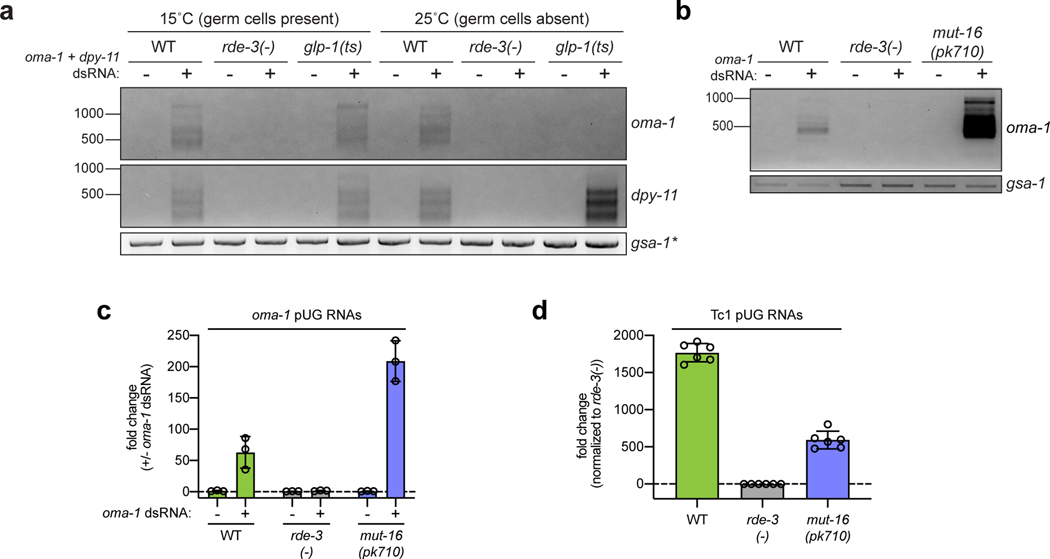
Extended Data Fig. 6. Mutator foci likely coordinate pUG RNA biogenesis within germ cells.
a,dpy-11 and oma-1 pUG PCR (Fig. 1a) were performed on total RNA from glp-1(q224/ts) animals grown at 15°C (germ cells present) or 25°C (<99% of germ cells), +/− oma-1 and dpy-11 dsRNA. Data is representative of 2 biologically independent experiments. *Note: the samples in a are the same as those used in Fig. 3e and, therefore, the gsa-1 loading control is the same. b, oma-1 pUG PCR was performed on total RNA extracted from wild-type, rde-3(−), and mut-16(pk710) animals, +/− oma-1 dsRNA. Data is representative of 4 biologically independent experiments. c, qRT-PCR was used to quantify levels of oma-1 pUG RNAs in wild-type, rde-3(−), and mut-16(pk710) animals, +/− oma-1 dsRNA. Data is represented as fold change in the levels of oma-1 pUG RNAs +/− oma-1 dsRNA (y-axis) for each strain (x-axis). n=3 biologically independent samples per treatment for each strain. Error bars: s.d. of the mean. d, qRT-PCR was used to quantify levels of Tc1 pUG RNAs in wild-type, rde-3(−), and mut-16(pk710) animals. Note: the RNA samples used for d are the same as those used in c, except that the data for +/− oma-1 dsRNA samples were pooled for each strain. n=6 biologically independent samples for each strain. Error bars: s.d. of the mean. The analyses in c and d showed that mut-16 mutant animals produced more oma-1, but fewer Tc1, pUG RNAs, than wild-type animals. The increased levels of oma-1 pUG RNAs in mut-16(pk710) animals was also suggested by the gel in b. Together, these data suggest that Mutator foci likely have an important role in coordinating pUG RNA biogenesis in germ cells, as pUG RNA levels become misregulated in mut-16(pk710) mutants.