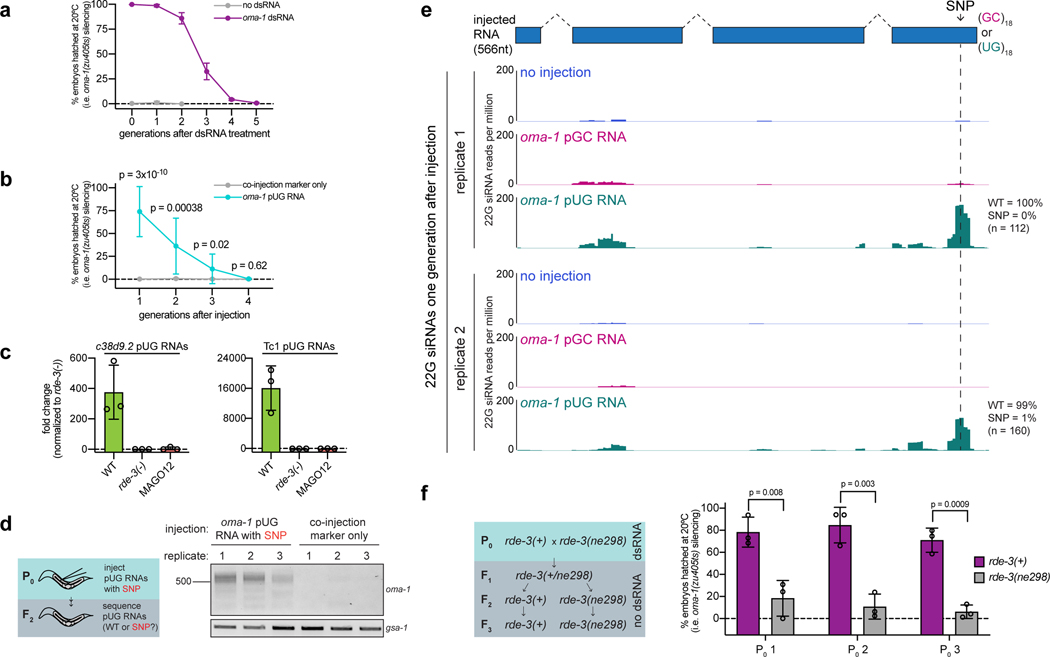
Extended Data Fig. 8. De novo pUGylation events in progeny are required for TEI.
a,oma-1(zu405ts) animals were fed bacteria expressing empty vector control or oma-1 dsRNA and % embryos hatched at 20°C was scored for 6 generations. Error bars represent s.d. of the mean of three biologically independent experiments. For each experiment, % embryos hatched at 20°C was averaged for 6 individual animals per treatment for each genotype. b, rde-1(ne219); oma-1(zu405ts) animals were injected with co-injection marker alone (n=12) or co-injection marker + oma-1 pUG RNA (n=19) and % embryos hatched at 20°C was scored for four generations in lineages of animals established from injected parents (see Methods for details of experimental setup). Error bars: s.d. of the mean. p-values: two-tailed unpaired Student’s t-test. c, c38d9.2 and Tc1 pUG RNA expression quantified in embryos harvested from wild-type, rde-3(−) or MAGO12 animals using qRT-PCR. Fold change normalized to rde-3(−). Each point (n) represents a biologically independent replicate, n=3 independent replicates/strain. Error bars: s.d. of the mean. d, Same experiment as Fig. 5d. rde-1(ne219); oma-1(zu405ts) animals were injected with an oma-1(SNP) pUG RNA or with co-injection marker only. Co-injection marker-expressing F1 progeny were picked and allowed to lay their F2 broods. oma-1 pUG PCR was performed on total RNA from F2 progeny. Shown is data from three biological replicates. e, Two biological replicates of small RNAs sequenced from the progeny of rde-1(ne219); oma-1(zu405ts) animals injected with oma-1(SNP) pUG or pGC RNAs are shown. Dotted line indicates the location of the SNP incorporated into oma-1. Distribution of 22G siRNAs mapping antisense to oma-1 is shown, with 22G siRNA reads normalized to reads per million total reads. In Fig. 4d and Extended Data Fig. 7a, small RNAs were sequenced 1–4 hours after injection and 100% of 22G siRNAs antisense to the region of the engineered SNP in oma-1 were found to encode the complement of the SNP. Shown here, <1% of 22G siRNAs from progeny of injected animals encoded the SNP complement. Note: siRNAs mapping near the pUG tail were observed only after oma-1(SNP) pUG RNA injection (pUG-specific siRNAs). For unknown reasons, both oma-1(SNP) pUG and pGC RNAs triggered small RNA production 5’ of the pUG-specific siRNAs. It is possible that these siRNAs were triggered by systems that respond to foreign RNAs, such as the piRNA system. Further work will be needed to ascertain the etiology of these siRNAs. f, Same experiment as Fig. 5e. oma-1(zu405ts) hermaphrodites were fed oma-1 dsRNA and crossed to rde-3(ne298); oma-1(zu405ts) males. F2 progeny from this cross were genotyped for rde-3(ne298). WT and rde-3(ne298) homozygous F3 progeny were phenotyped for % embryonic arrest at 20°C. 3 biologically independent crosses (P0 1–3) were performed. Error bars: s.d. of the mean. p-values: two-tailed unpaired Student’s t-tests.