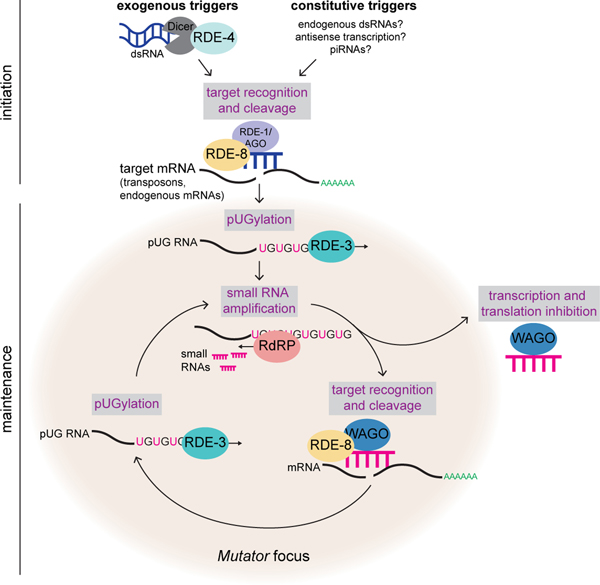
Extended Data Fig. 9. Working model for pUG RNA/siRNA cycling during RNAi.
Initiation: exogenous and constitutive (i.e. genomically encoded such as dsRNA, piRNAs) triggers direct RDE-3 to pUGylate RNAs previously fragmented by factors in the RNAi pathway. Maintenance: pUG RNAs are templates for 2° siRNA synthesis by RdRPs. Argonaute proteins (termed WAGOs) bind 2° siRNAs and: 1) target homologous RNAs for transcriptional and translational silencing29,34,66,67, as well as 2) direct the cleavage and de novo pUGylation of additional mRNAs. In this way, cycles of pUG RNA-based siRNA production and siRNA-directed mRNA pUGylation maintain silencing over time and across generations. This model shows germline perinuclear condensates termed Mutator foci as the likely sites of pUG RNA biogenesis in germ cells for several reasons. RDE-3 localizes to Mutator foci17 and we show in Fig. 3d that endogenous pUG RNAs localize to Mutator foci. The fact that enzyme and enzyme product both localize to Mutator foci suggests that Mutator foci may be sites of RNA pUGylation. In addition, while pUG RNAs are still made in mut-16 mutants (Extended Data Fig. 6b-d), which lack Mutator foci, the levels of both dsRNA-triggered and endogenous pUG RNAs are misregulated. Thus, while RDE-3 still has enzymatic activity in the absence of Mutator foci, these perinuclear condensates are likely coordinating target recognition and pUGylation in wild-type animals. Indeed, both the endonuclease RDE-8, which cleaves mRNAs targeted by dsRNA12, and the RdRP RRF-117 also localize to Mutator foci, further suggesting that pUG RNA/siRNA cycling occurs in Mutator foci.
Previous studies have shown that animals lacking RDE-3 still produce some 22G endo- siRNAs, including 22G siRNAs that associate with the Argonaute CSR-1 and whose biogenesis depends upon the RdRP EGO-126,68. Thus, EGO-1 may also produce some 22G siRNAs via a pUG RNA-independent mechanism.
A previous study showed that, in rrf-1 mutants that lack germlines, sel-1 RNAi causes a small fraction of sel-1 mRNA fragments to be uridylated in a largely RDE-3–dependent manner in the soma12. This data suggests that, in somatic tissues, RDE-3 may add non-templated Us to the 3’ termini of mRNA fragments generated during RNAi. It was proposed that this uridylation may be important for turnover or decay of RNAi targets12. Our work, combined with this earlier data about RDE-3–dependent uridylation12, suggests two models. First, RDE-3 may possess two distinct catalytic activities: uridylation and pUGylation. According to this model, RDE-3 might add Us or UGs depending on context (e.g. cell/tissue-type or developmental timing). Alternatively, the mRNA uridylation observed in the soma could depend upon RDE-3 and the pUGylation system, but may be mediated by another, currently unknown, poly(U) polymerase.