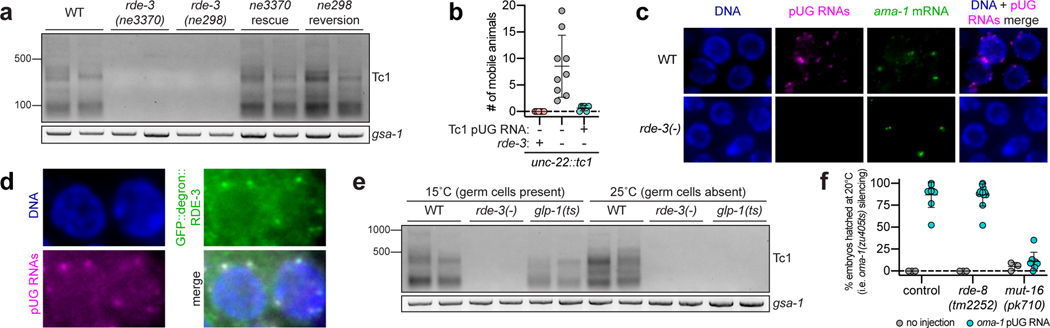
Figure 3. Endogenous pUG RNAs exist and localize to germline Mutator foci.
a, Tc1 pUG PCR (Fig. 1a) on total RNA from two replicates of indicated genotypes (rescue/reversion as in Fig. 1b). WT vs. rde-3(ne3370) and WT vs. rde-3(ne298) is representative of >5 and 2 biologically independent experiments, respectively. b, 18 and 22 rde-3(−); unc-22::tc1 animals were injected with Tc1 pUG RNA + co-injection marker or co-injection marker alone, respectively. Each data point (n) represents # of mobile progeny (indicating Tc1 mobilized from unc-22) laid by 25 randomly pooled co-injection marker–expressing progeny derived from injected animals (see Methods). n=9 for co-injection marker only, 6 for Tc1 pUG RNA + co-injection marker, 6 for noninjected rde-3(+); unc-22::tc1. Error bars: s.d. of the mean. c-d, Fluorescence micrographs of adult pachytene stage germ cell nuclei. DNA stained with 4’,6-diamidino-2-phenylindole (DAPI, blue). Data is representative of 3 biologically independent experiments. c, RNA FISH to detect pUG RNAs on germlines dissected from WT or rde-3(−) animals using (AC)9 DNA oligo conjugated to Alexa 647 (magenta). Positive control: RNA FISH to detect ama-1 mRNA (green). d, pUG RNA FISH (magenta) combined with immunofluorescence to detect GFP::degron::RDE-3 (green). e, Tc1 pUG PCR on total RNA isolated from replicates of glp-1(q224/ts) animals grown at 15°C (germ cells present) or 25°C (<99% of germ cells). Data is representative of 2 biologically independent experiments. f, Control, rde-8(tm2252) or mut-16(pk710) animals (all rde-1(ne219); oma-1(zu405ts) background) were injected with oma-1 pUG RNAs (n=9, 12 and 8, respectively) and % embryos hatched scored at 20°C. n=3 for all no injection. Error bars: s.d. of the mean.