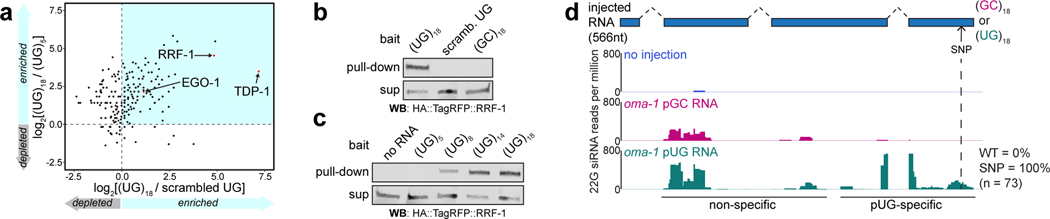
Figure 4. pUG RNAs are templates for RdRPs.
a, LC-MS/MS was performed on proteins that bound to 5’ biotinylated RNA oligos [(UG)5, (UG)18 or 36 scrambled UGs] conjugated to streptavidin beads. Shown is a scatter plot of log2-transformed fold enrichment in (UG)18 vs. scrambled UG pull-down (x-axis) and (UG)18 vs. (UG)5 pull-down (y-axis). Proteins enriched ≥2-fold in (UG)18 vs. beads-only pull-down are plotted. b-c, Indicated 5’ biotinylated RNA oligos were conjugated to streptavidin beads and incubated with extracts from animals expressing HA::tagRFP::RRF-1. Bead-bound material (pull-down) and supernatant (sup) were subjected to ɑ-HA immunoblotting. Data is representative of 2 biologically independent experiments. d, rde-1(ne219); oma-1(zu405ts) animals were injected with SNP-containing (dotted line) oma-1 (oma-1(SNP)) pUG or pGC-tailed RNAs and collected 1–4 hours later; small RNAs (20–30nts) were sequenced. Distribution of 22G siRNAs mapping antisense to oma-1 is shown, with 22G siRNA reads normalized to reads per million total reads. pUG-specific: 22G siRNAs observed only after oma-1(SNP) pUG RNA injection; non-specific: 22G siRNAs observed after oma-1(SNP) pUG and pGC RNA injections. See Extended Data Fig. 7 for biological replicate and details about sequenced small RNAs.