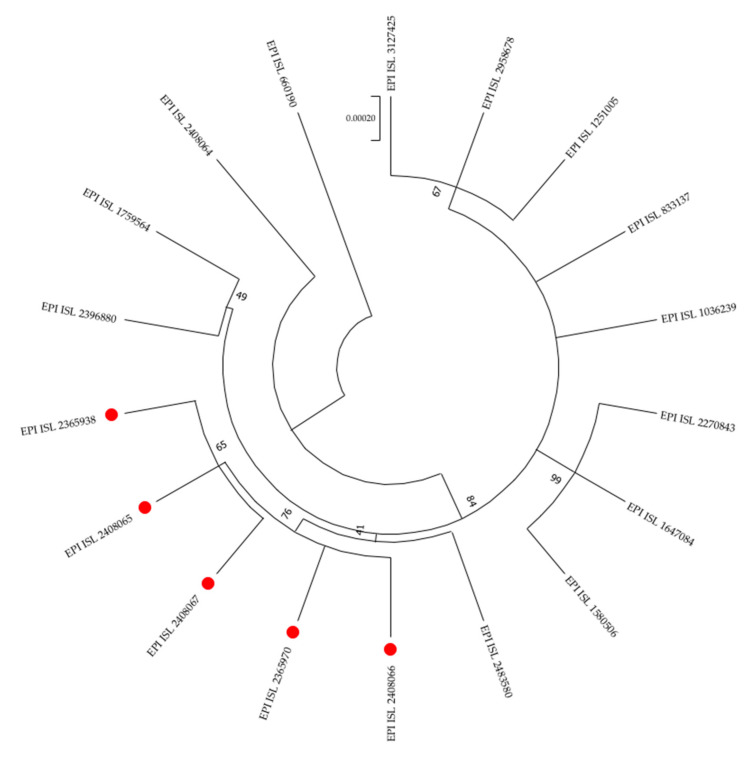
Figure 1.
Phylogenetic tree of 18 SARS-CoV-2 full genome sequences, including the five genomes examined in this study (red dots). The reference SARS-CoV-2 genome (GISAID accession number: EPI_ISL_660190) was included to root the tree. P.1 reference strain has been used to construct the tree (GISAID accession numbers: EPI_ISL_833137). Other Italian P.1 SARS-CoV-2 sequences have been used for comparison (GISAID accession numbers: EPI_ISL_2408064, EPI_ISL_1759564, EPI_ISL_2396880, EPI_ISL_2483580, EPI_ISL_1580506, EPI_ISL_1647084, EPI_ISL_2270843, EPI_ISL_1036239, EPI_ISL_1251005, EPI_ISL_2958678, EPI_ISL_3127425). Evolutionary analyses were conducted in MEGAX by using the maximum likelihood method and Tamura-Nei model. The robustness of branching pattern was tested by 1000 bootstrap replications. Bootstrap values are reported. The scale bar indicates nucleotides substitutions per site.