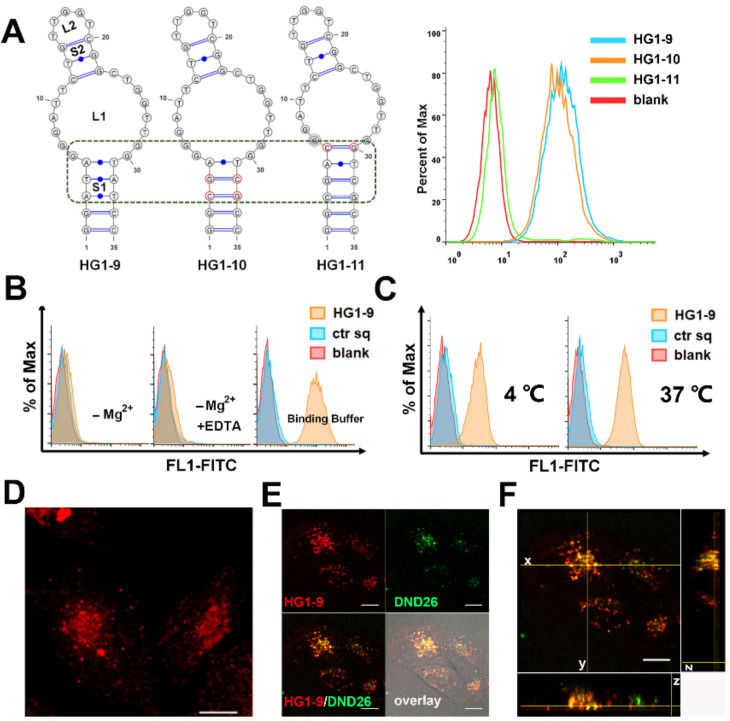
Figure 3.
Characterization and optimization of HG1-9. (A) The predicted secondary structures of HG1-9, and its mutated sequences HG1-10 and HG1-11; and flow cytometry analysis of their binding to Jurkat cells. (B,C) The binding conditions that affect HG1-9 binding to Jurkat cells. HG1-9 incubated with cells at 4 °C in different buffers: PBS with 5 mM MgCl2, PBS without MgCl2and PBS with 5 mM EDTA (B). HG1-9 incubated with cells at temperatures of 4 and 37 °C in the binding buffer (C). Blank represents Jurkat cells without any treatment, ctr sq was the negative control. (D) 2D projection of the maximum intensity from axial (Z) dimension of fluorescence of HG1-9 (10 nM, labeled by TAMRA) within HeLa cells after incubation for 4 h at 37 °C. (E) Confocal imaging of HeLa cells co-stained by HG1-9 (labeled by TAMRA) and green Lysotraker (DND-26) at 37 °C for 30 min. (F) 3D images of HeLa cells in (E), the images respectively represent x–y plane, x–z plane and y–z plane. The scale is 10 μm.